BrainMaps
 | |
|---|---|
| Content | |
| Description | Interactive zoomable high-resolution digital brain atlas |
| Data types captured | Neuroanatomy, Histology |
| Organisms | includes: Man, monkey, cat, mouse, opossum, goldfish, platypus, dog, owl, chicken, rat, and more |
| Access | |
| Website | http://brainmaps.org/ |
| Miscellaneous | |
| License | All image dataset is copyrighted to their respective owners, if none indicated, to the UC Regents Davis campus.[1] |

a: choosing from some hundreds of coronal sections.
b: certain coronal section shown.
c: zooming up of insular cortex region.
d: further zooming up of insular cortex. Nissl stained neurons are visible. This slice can be accessed through this link. [1]
BrainMaps is an NIH-funded interactive zoomable high-resolution digital brain atlas and virtual microscope that is based on more than 140 million megapixels (140 terabytes) of scanned images of serial sections of both primate and non-primate brains and that is integrated with a high-speed database for querying and retrieving data about brain structure and function over the internet.
Currently featured are complete brain atlas datasets for 16 species; a few of which are: Macaca mulatta, Chlorocebus aethiops, Felis silvestris catus, Mus musculus, Rattus norvegicus, and Tyto alba.
The project's principal investigator was UC Davis neuroscientist Ted Jones from 2005 through 2011, after which the role was taken by W. Martin Usrey.
Description[]
BrainMaps uses multiresolution image formats for representing massive brain images, and a dHTML/Javascript front-end user interface for image navigation, both similar to the way that Google Maps works for geospatial data.
BrainMaps is one of the most massive online neuroscience databases and image repositories and features the highest-resolution whole brain atlas ever constructed.[2][3]
Extensions to interactive 3-dimensional visualization have been developed through OpenGL-based desktop applications.[4] Freely available image analysis tools enable end-users to datamine online images at the sub-neuronal level. BrainMaps has been used in both research [5][6] and didactic settings.
Additional images[]

Massive brain images are formatted as multiresolution image pyramids, enabling rapid navigation by loading small image tiles instead of the entire image.
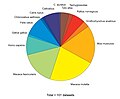
Datasets as a function of species at BrainMaps.
See also[]
References[]
- ^ Terms of Use, BrainMaps.org Access date: 2014-02-11
- ^ Mikula, S; Trotts I; Stone JM; Jones EG (2007). "Internet-Enabled High-Resolution Brain Mapping and Virtual Microscopy". NeuroImage. 35 (1): 9–15. doi:10.1016/j.neuroimage.2006.11.053. PMC 1890021. PMID 17229579.
- ^ Mikula, S; Stone JM; Jones EG (2008). "BrainMaps.org - Interactive High-Resolution Digital Brain Atlases and Virtual Microscopy". Brains Minds Media. 3: bmm1426. PMC 2614326. PMID 19129928.
- ^ Trotts, I; Mikula S; Jones EG (2007). "Interactive Visualization of Multiresolution Image Stacks in 3D". NeuroImage. 35 (3): 1038–43. doi:10.1016/j.neuroimage.2007.01.013. PMC 2492583. PMID 17336095.
- ^ Mikula, S; Manger PR; Jones EG (2007). "The thalamus of the monotremes: cyto- and myeloarchitecture and chemical neuroanatomy". Philos Trans R Soc Lond B Biol Sci. 363 (1502): 2415–40. doi:10.1098/rstb.2007.2133. PMC 2606803. PMID 17553780.
- ^ Mikula, S; Parrish SK; Trimmer JS; Jones EG (2009). "Complete 3-D visualization of primate striosomes by KChIP1 immunostaining". J Comp Neurol. 514 (5): 507–17. doi:10.1002/cne.22051. PMC 2737266. PMID 19350670.
External links[]
| Wikimedia Commons has media related to BrainMaps. |
- Online databases
- Anatomy websites
- Biological databases

