Diatom
| Diatoms Temporal range: Jurassic–Present
PreꞒ
Ꞓ
O
S
D
C
P
T
J
K
Pg
N
| |
|---|---|
| |
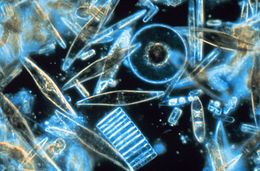
| Light microscopy of a sampling of marine diatoms found living between crystals of annual sea ice in Antarctica, showing a multiplicity of sizes and shapes | |
| Scientific classification | |
| Clade: | SAR |
| Phylum: | Ochrophyta |
| Subphylum: | Khakista |
| Class: | Bacillariophyceae Dangeard, 1933[1] |
| Synonyms | |
Diatoms (diá-tom-os 'cut in half', from diá, 'through' or 'apart', and the root of tém-n-ō, 'I cut')[6] are a major group of algae,[7] specifically microalgae, found in the oceans, waterways and soils of the world. Living diatoms make up a significant portion of the Earth's biomass: they generate about 20 to 50 percent of the oxygen produced on the planet each year,[8][9] take in over 6.7 billion metric tons of silicon each year from the waters in which they live,[10] and constitute nearly half of the organic material found in the oceans. The shells of dead diatoms can reach as much as a half-mile (800 m) deep on the ocean floor, and the entire Amazon basin is fertilized annually by 27 million tons of diatom shell dust transported by transatlantic winds from the African Sahara, much of it from the Bodélé Depression, which was once made up of a system of fresh-water lakes.[11][12]
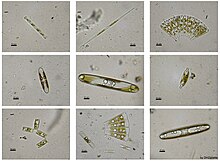
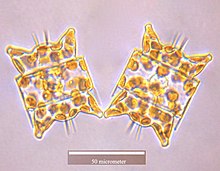
Diatoms are unicellular: they occur either as solitary cells or in colonies, which can take the shape of ribbons, fans, zigzags, or stars. Individual cells range in size from 2 to 200 micrometers.[13] In the presence of adequate nutrients and sunlight, an assemblage of living diatoms doubles approximately every 24 hours by asexual multiple fission; the maximum life span of individual cells is about six days.[14] Diatoms have two distinct shapes: a few (centric diatoms) are radially symmetric, while most (pennate diatoms) are broadly bilaterally symmetric. A unique feature of diatom anatomy is that they are surrounded by a cell wall made of silica (hydrated silicon dioxide), called a frustule.[15] These frustules have structural coloration due to their photonic nanostructure, prompting them to be described as "jewels of the sea" and "living opals". Movement in diatoms primarily occurs passively as a result of both water currents and wind-induced water turbulence; however, male gametes of centric diatoms have flagella, permitting active movement for seeking female gametes. Similar to plants, diatoms convert light energy to chemical energy by photosynthesis, although this shared autotrophy evolved independently in both lineages.
Unusually for autotrophic organisms, diatoms possess a urea cycle, a feature that they share with animals, although this cycle is used to different metabolic ends in diatoms. The family Rhopalodiaceae also possess a cyanobacterial endosymbiont called a spheroid body. This endosymbiont has lost its photosynthetic properties, but has kept its ability to perform nitrogen fixation, allowing the diatom to fix atmospheric nitrogen.[16] Other diatoms in symbiosis with nitrogen-fixing cyanobacteria are among the genera Hemiaulus, Rhizosolenia and Chaetoceros.[17]
The study of diatoms is a branch of phycology. Diatoms are classified as eukaryotes, organisms with a membrane-bound cell nucleus, that separates them from the prokaryotes archaea and bacteria. Diatoms are a type of plankton called phytoplankton, the most common of the plankton types. Diatoms also grow attached to benthic substrates, floating debris, and on macrophytes. They comprise an integral component of the periphyton community.[18] Another classification divides plankton into eight types based on size: in this scheme, diatoms are classed as microalgae. Several systems for classifying the individual diatom species exist.
Fossil evidence suggests that diatoms originated during or before the early Jurassic period, which was about 150 to 200 million years ago. The oldest fossil evidence for diatoms is a specimen of extant genus in Late Jurassic aged amber from Thailand.[19]
Diatoms are used to monitor past and present environmental conditions, and are commonly used in studies of water quality. Diatomaceous earth (diatomite) is a collection of diatom shells found in the earth's crust. They are soft, silica-containing sedimentary rocks which are easily crumbled into a fine powder and typically have a particle size of 10 to 200 μm. Diatomaceous earth is used for a variety of purposes including for water filtration, as a mild abrasive, in cat litter, and as a dynamite stabilizer.
Displays overlays from four fluorescent channels
(b) Cyan: [PLL-A546 fluorescence] - generic counterstain for visualising eukaryotic cell surfaces
(c) Blue: [Hoechst fluorescence] - stains DNA, identifies nuclei
(d) Red: [chlorophyll autofluorescence] - resolves chloroplasts [20]
Morphology[]
Diatoms are generally 2 to 200 micrometers in size,[13] with a few larger species. Their yellowish-brown chloroplasts, the site of photosynthesis, are typical of heterokonts, having four membranes and containing pigments such as the carotenoid fucoxanthin. Individuals usually lack flagella, but they are present in male gametes of the centric diatoms and have the usual heterokont structure, including the hairs (mastigonemes) characteristic in other groups.
Diatoms are often referred as "jewels of the sea" or "living opals" due to their optical properties.[23] The biological function of this structural coloration is not clear, but it is speculated that it may be related to communication, camouflage, thermal exchange and/or UV protection.[24]
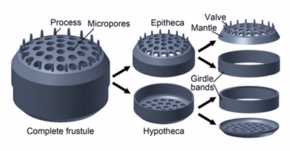
Diatoms build intricate hard but porous cell walls called frustules composed primarily of silica.[25]:25–30 This siliceous wall[26] can be highly patterned with a variety of pores, ribs, minute spines, marginal ridges and elevations; all of which can be used to delineate genera and species.
The cell itself consists of two halves, each containing an essentially flat plate, or valve, and marginal connecting, or girdle band. One half, the hypotheca, is slightly smaller than the other half, the epitheca. Diatom morphology varies. Although the shape of the cell is typically circular, some cells may be triangular, square, or elliptical. Their distinguishing feature is a hard mineral shell or frustule composed of opal (hydrated, polymerized silicic acid).

2) Nucleolus; Location of the chromosomes
3) Golgi complex; modifies proteins and sends them out of the cell
4) Cell Wall; Outer membrane of the cell
5) Pyrenoid; center of carbon fixation
6) Chromatophore; pigment carrying membrane structure
7) Vacuoles; vesicle of a cell that contains fluid bound by a membrane
8) Cytoplasmic strands; hold the nucleus
9) Mitochondria; create ATP (energy) for the cell
10) Valves/Striae; allow nutrients in, and waste out, of the cell
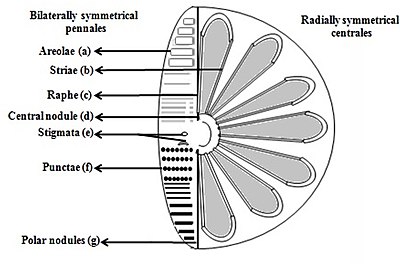
b) Striae (pores, punctae, spots or dots in a line on the surface)
c) Raphe (slit in the valves)
d) Central nodule (thickening of wall at the midpoint of raphe)
e) Stigmata (holes through valve surface which looks rounded externally but with a slit like internal)
f) Punctae (spots or small perforations on the surface)
g) Polar nodules (thickening of wall at the distal ends of the raphe) [27][28]

Diatoms are divided into two groups that are distinguished by the shape of the frustule: the centric diatoms and the pennate diatoms.
Pennate diatoms are bilaterally symmetric. Each one of their valves have openings that are slits along the raphes and their shells are typically elongated parallel to these raphes. They generate cell movement through cytoplasm that streams along the raphes, always moving along solid surfaces.
Centric diatoms are radially symmetric. They are composed of upper and lower valves – epitheca and hypotheca – each consisting of a valve and a girdle band that can easily slide underneath each other and expand to increase cell content over the diatoms progression. The cytoplasm of the centric diatom is located along the inner surface of the shell and provides a hollow lining around the large vacuole located in the center of the cell. This large, central vacuole is filled by a fluid known as "cell sap" which is similar to seawater but varies with specific ion content. The cytoplasmic layer is home to several organelles, like the chloroplasts and mitochondria. Before the centric diatom begins to expand, its nucleus is at the center of one of the valves and begins to move towards the center of the cytoplasmic layer before division is complete. Centric diatoms have a variety of shapes and sizes, depending on from which axis the shell extends, and if spines are present.

Behaviour[]
Most centric and araphid pennate diatoms are nonmotile, and their relatively dense cell walls cause them to readily sink. Planktonic forms in open water usually rely on turbulent mixing of the upper layers of the oceanic waters by the wind to keep them suspended in sunlit surface waters. Many planktonic diatoms have also evolved features that slow their sinking rate, such as spines or the ability to grow in colonial chains.[30] These adaptations increase their surface area to volume ratio and drag, allowing them to stay suspended in the water column longer. Individual cells may regulate buoyancy via an ionic pump.[31]
Some pennate diatoms are capable of a type of locomotion called "gliding", which allows them to move across surfaces via adhesive mucilage secreted through the raphe (an elongated slit in the valve face).[32][33] In order for a diatom cell to glide, it must have a solid substrate for the mucilage to adhere to.
Cells are solitary or united into colonies of various kinds, which may be linked by siliceous structures; mucilage pads, stalks or tubes; amorphous masses of mucilage; or by threads of chitin (polysaccharide), which are secreted through strutted processes of the cell.

Gran, 1897



This projection of a stack of confocal images shows the diatoms' cell wall (cyan), chloroplasts (red), DNA (blue), membranes and organelles (green).
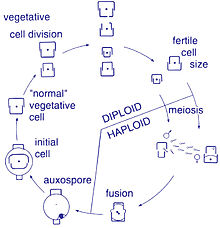
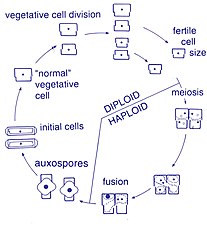
Life cycle[]
Reproduction and cell size[]
Reproduction among these organisms is asexual by binary fission, during which the diatom divides into two parts, producing two "new" diatoms with identical genes. Each new organism receives one of the two frustules – one larger, the other smaller – possessed by the parent, which is now called the epitheca; and is used to construct a second, smaller frustule, the hypotheca. The diatom that received the larger frustule becomes the same size as its parent, but the diatom that received the smaller frustule remains smaller than its parent. This causes the average cell size of this diatom population to decrease.[13] It has been observed, however, that certain taxa have the ability to divide without causing a reduction in cell size.[34] Nonetheless, in order to restore the cell size of a diatom population for those that do endure size reduction, sexual reproduction and auxospore formation must occur.[13]
Cell division[]
Vegetative cells of diatoms are diploid (2N) and so meiosis can take place, producing male and female gametes which then fuse to form the zygote. The zygote sheds its silica theca and grows into a large sphere covered by an organic membrane, the auxospore. A new diatom cell of maximum size, the initial cell, forms within the auxospore thus beginning a new generation. Resting spores may also be formed as a response to unfavourable environmental conditions with germination occurring when conditions improve.[25]
Sperm motility[]
Diatoms are mostly non-motile; however, sperm found in some species can be flagellated, though motility is usually limited to a gliding motion.[25] In centric diatoms, the small male gametes have one flagellum while the female gametes are large and non-motile (oogamous). Conversely, in pennate diatoms both gametes lack flagella (isogamous).[13] Certain araphid species, that is pennate diatoms without a raphe (seam), have been documented as anisogamous and are, therefore, considered to represent a transitional stage between centric and raphid pennate diatoms, diatoms with a raphe.[34]
Degradation by microbes[]
Certain species of bacteria in oceans and lakes can accelerate the rate of dissolution of silica in dead and living diatoms by using hydrolytic enzymes to break down the organic algal material.[35][36]
Ecology[]

Fluxes are in T mol Si y−1 (28 million metric tons of silicon per year)


versus silicate concentration [39]
Distribution[]
Diatoms are a widespread group and can be found in the oceans, in fresh water, in soils, and on damp surfaces. They are one of the dominant components of phytoplankton in nutrient-rich coastal waters and during oceanic spring blooms, since they can divide more rapidly than other groups of phytoplankton.[40] Most live pelagically in open water, although some live as surface films at the water-sediment interface (benthic), or even under damp atmospheric conditions. They are especially important in oceans, where they contribute an estimated 45% of the total oceanic primary production of organic material.[41] Spatial distribution of marine phytoplankton species is restricted both horizontally and vertically.[42][25]
Growth[]
Planktonic diatoms in freshwater and marine environments typically exhibit a "boom and bust" (or "bloom and bust") lifestyle. When conditions in the upper mixed layer (nutrients and light) are favourable (as at the spring), their competitive edge and rapid growth rate[40] enables them to dominate phytoplankton communities ("boom" or "bloom"). As such they are often classed as opportunistic r-strategists (i.e. those organisms whose ecology is defined by a high growth rate, r).
Contribution to modern oceanic silicon cycle[]
Diatoms contribute in a significant way to the modern oceanic silicon cycle: they are the source of the vast majority of biological production.
Impact[]
The freshwater diatom Didymosphenia geminata, commonly known as Didymo, causes severe environmental degradation in water-courses where it blooms, producing large quantities of a brown jelly-like material called "brown snot" or "rock snot". This diatom is native to Europe and is an invasive species both in the antipodes and in parts of North America.[43][44] The problem is most frequently recorded from Australia and New Zealand.[45]
When conditions turn unfavourable, usually upon depletion of nutrients, diatom cells typically increase in sinking rate and exit the upper mixed layer ("bust"). This sinking is induced by either a loss of buoyancy control, the synthesis of mucilage that sticks diatoms cells together, or the production of heavy resting spores. Sinking out of the upper mixed layer removes diatoms from conditions unfavourable to growth, including grazer populations and higher temperatures (which would otherwise increase cell metabolism). Cells reaching deeper water or the shallow seafloor can then rest until conditions become more favourable again. In the open ocean, many sinking cells are lost to the deep, but refuge populations can persist near the thermocline.
Ultimately, diatom cells in these resting populations re-enter the upper mixed layer when vertical mixing entrains them. In most circumstances, this mixing also replenishes nutrients in the upper mixed layer, setting the scene for the next round of diatom blooms. In the open ocean (away from areas of continuous upwelling[46]), this cycle of bloom, bust, then return to pre-bloom conditions typically occurs over an annual cycle, with diatoms only being prevalent during the spring and early summer. In some locations, however, an autumn bloom may occur, caused by the breakdown of summer stratification and the entrainment of nutrients while light levels are still sufficient for growth. Since vertical mixing is increasing, and light levels are falling as winter approaches, these blooms are smaller and shorter-lived than their spring equivalents.
In the open ocean, the diatom (spring) bloom is typically ended by a shortage of silicon. Unlike other minerals, the requirement for silicon is unique to diatoms and it is not regenerated in the plankton ecosystem as efficiently as, for instance, nitrogen or phosphorus nutrients. This can be seen in maps of surface nutrient concentrations – as nutrients decline along gradients, silicon is usually the first to be exhausted (followed normally by nitrogen then phosphorus).
Because of this bloom-and-bust cycle, diatoms are believed to play a disproportionately important role in the export of carbon from oceanic surface waters[46][47] (see also the biological pump). Significantly, they also play a key role in the regulation of the biogeochemical cycle of silicon in the modern ocean.[41][37]
Reason for success[]
Diatoms are ecologically successful, and occur in virtually every environment that contains water – not only oceans, seas, lakes, and streams, but also soil and wetlands.[citation needed] The use of silicon by diatoms is believed by many researchers to be the key to this ecological success. Raven (1983)[48] noted that, relative to organic cell walls, silica frustules require less energy to synthesize (approximately 8% of a comparable organic wall), potentially a significant saving on the overall cell energy budget. In a now classic study, Egge and Aksnes (1992)[39] found that diatom dominance of mesocosm communities was directly related to the availability of silicic acid – when concentrations were greater than 2 μmol m−3, they found that diatoms typically represented more than 70% of the phytoplankton community. Other researchers[49] have suggested that the biogenic silica in diatom cell walls acts as an effective pH buffering agent, facilitating the conversion of bicarbonate to dissolved CO2 (which is more readily assimilated). More generally, notwithstanding these possible advantages conferred by their use of silicon, diatoms typically have higher growth rates than other algae of the same corresponding size.[40]
Sources for collection[]
Diatoms can be obtained from multiple sources.[50] Marine diatoms can be collected by direct water sampling, and benthic forms can be secured by scraping barnacles, oyster and other shells. Diatoms are frequently present as a brown, slippery coating on submerged stones and sticks, and may be seen to "stream" with river current. The surface mud of a pond, ditch, or lagoon will almost always yield some diatoms. Living diatoms are often found clinging in great numbers to filamentous algae, or forming gelatinous masses on various submerged plants. Cladophora is frequently covered with Cocconeis, an elliptically shaped diatom; Vaucheria is often covered with small forms. Since diatoms form an important part of the food of molluscs, tunicates, and fishes, the alimentary tracts of these animals often yield forms that are not easily secured in other ways. Diatoms can be made to emerge by filling a jar with water and mud, wrapping it in black paper and letting direct sunlight fall on the surface of the water. Within a day, the diatoms will come to the top in a scum and can be isolated.[50]
Biochemistry[]

| Part of a series on |
| Plankton |
|---|
 |
Energy source[]
Diatoms are mainly photosynthetic; however a few are obligate heterotrophs and can live in the absence of light provided an appropriate organic carbon source is available.[51][52]
Silica metabolism[]
Diatom cells are contained within a unique silica cell wall known as a frustule made up of two valves called thecae, that typically overlap one another.[53] The biogenic silica composing the cell wall is synthesised intracellularly by the polymerisation of silicic acid monomers. This material is then extruded to the cell exterior and added to the wall. In most species, when a diatom divides to produce two daughter cells, each cell keeps one of the two-halves and grows a smaller half within it. As a result, after each division cycle, the average size of diatom cells in the population gets smaller. Once such cells reach a certain minimum size, rather than simply divide, they reverse this decline by forming an auxospore. This expands in size to give rise to a much larger cell, which then returns to size-diminishing divisions.[citation needed] Auxospore production is almost always linked to meiosis and sexual reproduction.
The exact mechanism of transferring silica absorbed by the diatom to the cell wall is unknown. Much of the sequencing of diatom genes comes from the search for the mechanism of silica uptake and deposition in nano-scale patterns in the frustule. The most success in this area has come from two species, Thalassiosira pseudonana, which has become the model species, as the whole genome was sequenced and methods for genetic control were established, and Cylindrotheca fusiformis, in which the important silica deposition proteins silaffins were first discovered.[54] Silaffins, sets of polycationic peptides, were found in C. fusiformis cell walls and can generate intricate silica structures. These structures demonstrated pores of sizes characteristic to diatom patterns. When T. pseudonana underwent genome analysis it was found that it encoded a urea cycle, including a higher number of polyamines than most genomes, as well as three distinct silica transport genes.[55] In a phylogenetic study on silica transport genes from 8 diverse groups of diatoms, silica transport was found to generally group with species.[54] This study also found structural differences between the silica transporters of pennate (bilateral symmetry) and centric (radial symmetry) diatoms. The sequences compared in this study were used to create a diverse background in order to identify residues that differentiate function in the silica deposition process. Additionally, the same study found that a number of the regions were conserved within species, likely the base structure of silica transport.
These silica transport proteins are unique to diatoms, with no homologs found in other species, such as sponges or rice. The divergence of these silica transport genes is also indicative of the structure of the protein evolving from two repeated units composed of five membrane bound segments, which indicates either gene duplication or dimerization.[54] The silica deposition that takes place from the membrane bound vesicle in diatoms has been hypothesized to be a result of the activity of silaffins and long chain polyamines. This Silica Deposition Vesicle (SDV) has been characterized as an acidic compartment fused with Golgi-derived vesicles.[56] These two protein structures have been shown to create sheets of patterned silica in-vivo with irregular pores on the scale of diatom frustules. One hypothesis as to how these proteins work to create complex structure is that residues are conserved within the SDV's, which is unfortunately difficult to identify or observe due to the limited number of diverse sequences available. Though the exact mechanism of the highly uniform deposition of silica is as yet unknown, the Thalassiosira pseudonana genes linked to silaffins are being looked to as targets for genetic control of nanoscale silica deposition.
Urea cycle[]
A feature of diatoms is the urea cycle, which links them evolutionarily to animals. This was discovered in research carried out by Andrew Allen, Chris Bowler and colleagues. Their findings, published in 2011, that diatoms have a functioning urea cycle was highly significant, since prior to this, the urea cycle was thought to have originated with the metazoans which appeared several hundreds of millions of years before the diatoms. Their study showed that while diatoms and animals use the urea cycle for different ends, they are seen to be evolutionarily linked in such a way that animals and plants are not.[57]
Pigments[]
Major pigments of diatoms are chlorophylls a and c, beta-carotene, fucoxanthin, diatoxanthin and diadinoxanthin.[13]
Storage products[]
Storage products are chrysolaminarin and lipids.[25]
Taxonomy[]



Stephanodiscus hantzschii

Isthmia nervosaIsthmia nervosa
Odontella aurita
Diatoms belong to a large group of protists, many of which contain plastids rich in chlorophylls a and c. The group has been variously referred to as heterokonts, chrysophytes, chromists or stramenopiles. Many are autotrophs such as golden algae and kelp; and heterotrophs such as water moulds, opalinids, and actinophryid heliozoa. The classification of this area of protists is still unsettled. In terms of rank, they have been treated as a division, phylum, kingdom, or something intermediate to those. Consequently, diatoms are ranked anywhere from a class, usually called Diatomophyceae or Bacillariophyceae, to a division (=phylum), usually called Bacillariophyta, with corresponding changes in the ranks of their subgroups.
Genera and species[]
An estimated 20,000 extant diatom species are believed to exist, of which around 12,000 have been named to date according to Guiry, 2012[58] (other sources give a wider range of estimates[13][59][60][61]). Around 1,000–1,300 diatom genera have been described, both extant and fossil,[62][63] of which some 250–300 exist only as fossils.[64]
Classes and orders[]
For many years the diatoms—treated either as a class (Bacillariophyceae) or a phylum (Bacillariophyta)—were divided into just 2 orders, corresponding to the centric and the pennate diatoms (Centrales and Pennales). This classification was extensively overhauled by Round, Crawford and Mann in 1990 who treated the diatoms at a higher rank (division, corresponding to phylum in zoological classification), and promoted the major classification units to classes, maintaining the centric diatoms as a single class Coscinodiscophyceae, but splitting the former pennate diatoms into 2 separate classes, Fragilariophyceae and Bacillariophyceae (the latter older name retained but with an emended definition), between them encompassing 45 orders, the majority of them new.
Today (writing at mid 2020) it is recognised that the 1990 system of Round et al. is in need of revision with the advent of newer molecular work, however the best system to replace it is unclear, and current systems in widespread use such as AlgaeBase, the World Register of Marine Species and its contributing database , and the system for "all life" represented in Ruggiero et al., 2015, all retain the Round et al. treatment as their basis, albeit with diatoms as a whole treated as a class rather than division/phylum, and Round et al.'s classes reduced to subclasses, for better agreement with the treatment of phylogenetically adjacent groups and their containing taxa. (For references refer the individual sections below).
One proposal, by Linda Medlin and co-workers commencing in 2004, is for some of the centric diatom orders considered more closely related to the pennates to be split off as a new class, Mediophyceae, itself more closely aligned with the pennate diatoms than the remaining centrics. This hypothesis—later designated the Coscinodiscophyceae-Mediophyceae-Bacillariophyceae, or Coscinodiscophyceae+(Mediophyceae+Bacillariophyceae) (CMB) hypothesis—has been accepted by D.G. Mann among others, who uses it as the basis for the classification of diatoms as presented in Adl. et al.'s series of syntheses (2005, 2012, 2019), and also in the Bacillariophyta chapter of the 2017 Handbook of the Protists edited by Archibald et al., with some modifications reflecting the apparent non-monophyly of Medlin et al. original "Coscinodiscophyceae". Meanwhile, a group led by E.C. Theriot favours a different hypothesis of phylogeny, which has been termed the structural gradation hypothesis (SGH) and does not recognise the Mediophyceae as a monophyletic group, while another analysis, that of Parks et al., 2018, finds that the radial centric diatoms (Medlin et al.'s Coscinodiscophyceae) are not monophyletic, but supports the monophyly of Mediophyceae minus Attheya, which is an anomalous genus. Discussion of the relative merits of these conflicting schemes continues by the various parties involved.[65][66][67][68]
Adl et al., 2019 treatment[]
In 2019, Adl et al.[69] presented the following classification of diatoms, while noting: "This revision reflects numerous advances in the phylogeny of the diatoms over the last decade. Due to our poor taxon sampling outside of the Mediophyceae and pennate diatoms, and the known and anticipated diversity of all diatoms, many clades appear at a high classification level (and the higher level classification is rather flat)." This classification treats diatoms as a phylum (Diatomeae/Bacillariophyta), accepts the class Mediophyceae of Medlin and co-workers, introduces new subphyla and classes for a number of otherwise isolated genera, and re-ranks a number of previously established taxa as subclasses, but does not list orders or families. Inferred ranks have been added for clarity (Adl. et al. do not use ranks, but the intended ones in this portion of the classification are apparent from the choice of endings used, within the system of botanical nomenclature employed).
- Clade Derelle et al. 2016, emend. Cavalier-Smith 2017 (diatoms plus a subset of other ochrophyte groups)
- Phylum Diatomeae Dumortier 1821 [= Bacillariophyta Haeckel 1878] (diatoms)
- Subphylum Leptocylindrophytina D.G. Mann in Adl et al. 2019
- Class D.G. Mann in Adl et al. 2019 (Leptocylindrus, )
- Class D.G. Mann in Adl et al. 2019 ()
- Subphylum Ellerbeckiophytina D.G. Mann in Adl et al. 2019 (Ellerbeckia)
- Subphylum Probosciophytina D.G. Mann in Adl et al. 2019 ()
- Subphylum Melosirophytina D.G. Mann in Adl et al. 2019 (Aulacoseira, Melosira, Hyalodiscus, , , )
- Subphylum Coscinodiscophytina Medlin & Kaczmarska 2004, emend. (Actinoptychus, Coscinodiscus, Actinocyclus, , , )
- Subphylum Rhizosoleniophytina D.G. Mann in Adl et al. 2019 (Guinardia, , )
- Subphylum Arachnoidiscophytina D.G. Mann in Adl et al. 2019 ()
- Subphylum Bacillariophytina Medlin & Kaczmarska 2004, emend.
- Class Jouse & Proshkina-Lavrenko in Medlin & Kaczmarska 2004
- Subclass Chaetocerotophycidae Round & R.M. Crawford in Round et al. 1990, emend.
- Subclass Lithodesmiophycidae Round & R.M. Crawford in Round et al. 1990, emend.
- Subclass Thalassiosirophycidae Round & R.M. Crawford in Round et al. 1990
- Subclass Cymatosirophycidae Round & R.M. Crawford in Round et al. 1990
- Subclass Odontellophycidae D.G. Mann in Adl et al. 2019
- Subclass Chrysanthemodiscophycidae D.G. Mann in Adl et al. 2019
- Class D.G. Mann in Adl et al. 2019
- Subclass Biddulphiophycidae Round and R.M. Crawford in Round et al. 1990, emend.
- Biddulphiophyceae incertae sedis (Attheya)
- Class Bacillariophyceae Haeckel 1878, emend.
- Bacillariophyceae incertae sedis (Striatellaceae)
- Subclass Urneidophycidae Medlin 2016
- Subclass Fragilariophycidae Round in Round, Crawford & Mann 1990, emend.
- Subclass Bacillariophycidae D.G. Mann in Round, Crawford & Mann 1990, emend.
See taxonomy of diatoms for more details.
Gallery[]
- Scanning electron microscope images

Diatom Surirella spiralis

Diatoms Thalassiosira sp. on a membrane filter, pore size 0.4 μm.

Diatom Paralia sulcata.

Diatom Achanthes trinodis
Evolution and fossil record[]
Origin[]
Heterokont chloroplasts appear to derive from those of red algae, rather than directly from prokaryotes as occurred in plants. This suggests they had a more recent origin than many other algae. However, fossil evidence is scant, and only with the evolution of the diatoms themselves do the heterokonts make a serious impression on the fossil record.
Earliest fossils[]
The earliest known fossil diatoms date from the early Jurassic (~185 Ma ago),[70] although the molecular clock[70] and sedimentary[71] evidence suggests an earlier origin. It has been suggested that their origin may be related to the end-Permian mass extinction (~250 Ma), after which many marine niches were opened.[72] The gap between this event and the time that fossil diatoms first appear may indicate a period when diatoms were unsilicified and their evolution was cryptic.[73] Since the advent of silicification, diatoms have made a significant impression on the fossil record, with major fossil deposits found as far back as the early Cretaceous, and with some rocks such as diatomaceous earth, being composed almost entirely of them.
Relation to silicon cycle[]
Although diatoms may have existed since the Triassic, the timing of their ascendancy and "take-over" of the silicon cycle occurred more recently. Prior to the Phanerozoic (before 544 Ma), it is believed that microbial or inorganic processes weakly regulated the ocean's silicon cycle.[74][75][76] Subsequently, the cycle appears dominated (and more strongly regulated) by the radiolarians and siliceous sponges, the former as zooplankton, the latter as sedentary filter-feeders primarily on the continental shelves.[77] Within the last 100 My, it is thought that the silicon cycle has come under even tighter control, and that this derives from the ecological ascendancy of the diatoms.
However, the precise timing of the "take-over" remains unclear, and different authors have conflicting interpretations of the fossil record. Some evidence, such as the displacement of siliceous sponges from the shelves,[78] suggests that this takeover began in the Cretaceous (146 Ma to 66 Ma), while evidence from radiolarians suggests "take-over" did not begin until the Cenozoic (66 Ma to present).[79]
Relation to grasslands[]
The expansion of grassland biomes and the evolutionary radiation of grasses during the Miocene is believed to have increased the flux of soluble silicon to the oceans, and it has been argued that this promoted the diatoms during the Cenozoic era.[80][81] Recent work suggests that diatom success is decoupled from the evolution of grasses, although both diatom and grassland diversity increased strongly from the middle Miocene.[82]
Relation to climate[]
Diatom diversity over the Cenozoic has been very sensitive to global temperature, particularly to the equator-pole temperature gradient. Warmer oceans, particularly warmer polar regions, have in the past been shown to have had substantially lower diatom diversity. Future warm oceans with enhanced polar warming, as projected in global-warming scenarios,[83] could thus in theory result in a significant loss of diatom diversity, although from current knowledge it is impossible to say if this would occur rapidly or only over many tens of thousands of years.[82]
Method of investigation[]
The fossil record of diatoms has largely been established through the recovery of their siliceous frustules in marine and non-marine sediments. Although diatoms have both a marine and non-marine stratigraphic record, diatom biostratigraphy, which is based on time-constrained evolutionary originations and extinctions of unique taxa, is only well developed and widely applicable in marine systems. The duration of diatom species ranges have been documented through the study of ocean cores and rock sequences exposed on land.[84] Where diatom biozones are well established and calibrated to the geomagnetic polarity time scale (e.g., Southern Ocean, North Pacific, eastern equatorial Pacific), diatom-based age estimates may be resolved to within <100,000 years, although typical age resolution for Cenozoic diatom assemblages is several hundred thousand years.
Diatoms preserved in lake sediments are widely used for paleoenvironmental reconstructions of Quaternary climate, especially for closed-basin lakes which experience fluctuations in water depth and salinity.
Diversification[]
The Cretaceous record of diatoms is limited, but recent studies reveal a progressive diversification of diatom types. The Cretaceous–Paleogene extinction event, which in the oceans dramatically affected organisms with calcareous skeletons, appears to have had relatively little impact on diatom evolution.[85]
Turnover[]
Although no mass extinctions of marine diatoms have been observed during the Cenozoic, times of relatively rapid evolutionary turnover in marine diatom species assemblages occurred near the Paleocene–Eocene boundary,[86] and at the Eocene–Oligocene boundary.[87] Further turnover of assemblages took place at various times between the middle Miocene and late Pliocene,[88] in response to progressive cooling of polar regions and the development of more endemic diatom assemblages.
A global trend toward more delicate diatom frustules has been noted from the Oligocene to the Quaternary.[84] This coincides with an increasingly more vigorous circulation of the ocean's surface and deep waters brought about by increasing latitudinal thermal gradients at the onset of major ice sheet expansion on Antarctica and progressive cooling through the Neogene and Quaternary towards a bipolar glaciated world. This caused diatoms to take in less silica for the formation of their frustules. Increased mixing of the oceans renews silica and other nutrients necessary for diatom growth in surface waters, especially in regions of coastal and oceanic upwelling.
Genetics[]
Expressed sequence tagging[]
In 2002, the first insights into the properties of the Phaeodactylum tricornutum gene repertoire were described using 1,000 expressed sequence tags (ESTs).[89] Subsequently, the number of ESTs was extended to 12,000 and the diatom EST database was constructed for functional analyses.[90] These sequences have been used to make a comparative analysis between P. tricornutum and the putative complete proteomes from the green alga Chlamydomonas reinhardtii, the red alga Cyanidioschyzon merolae, and the diatom Thalassiosira pseudonana.[91] The diatom EST database now consists of over 200,000 ESTs from P. tricornutum (16 libraries) and T. pseudonana (7 libraries) cells grown in a range of different conditions, many of which correspond to different abiotic stresses.[92]
Genome sequencing[]
In 2004, the entire genome of the centric diatom, Thalassiosira pseudonana (32.4 Mb) was sequenced,[93] followed in 2008 with the sequencing of the pennate diatom, Phaeodactylum tricornutum (27.4 Mb).[94] Comparisons of the two reveal that the P. tricornutum genome includes fewer genes (10,402 opposed to 11,776) than T. pseudonana; no major synteny (gene order) could be detected between the two genomes. T. pseudonana genes show an average of ~1.52 introns per gene as opposed to 0.79 in P. tricornutum, suggesting recent widespread intron gain in the centric diatom.[94][95] Despite relatively recent evolutionary divergence (90 million years), the extent of molecular divergence between centrics and pennates indicates rapid evolutionary rates within the Bacillariophyceae compared to other eukaryotic groups.[94] Comparative genomics also established that a specific class of transposable elements, the Diatom Copia-like retrotransposons (or CoDis), has been significantly amplified in the P. tricornutum genome with respect to T. pseudonana, constituting 5.8 and 1% of the respective genomes.[96]
Endosymbiotic gene transfer[]
Diatom genomics brought much information about the extent and dynamics of the endosymbiotic gene transfer (EGT) process. Comparison of the T. pseudonana proteins with homologs in other organisms suggested that hundreds have their closest homologs in the Plantae lineage. EGT towards diatom genomes can be illustrated by the fact that the T. pseudonana genome encodes six proteins which are most closely related to genes encoded by the Guillardia theta (cryptomonad) nucleomorph genome. Four of these genes are also found in red algal plastid genomes, thus demonstrating successive EGT from red algal plastid to red algal nucleus (nucleomorph) to heterokont host nucleus.[93] More recent phylogenomic analyses of diatom proteomes provided evidence for a prasinophyte-like endosymbiont in the common ancestor of chromalveolates as supported by the fact the 70% of diatom genes of Plantae origin are of green lineage provenance and that such genes are also found in the genome of other stramenopiles. Therefore, it was proposed that chromalveolates are the product of serial secondary endosymbiosis first with a green algae, followed by a second one with a red algae that conserved the genomic footprints of the previous but displaced the green plastid.[97] However, phylogenomic analyses of diatom proteomes and chromalveolate evolutionary history will likely take advantage of complementary genomic data from under-sequenced lineages such as red algae.
Horizontal gene transfer[]
In addition to EGT, horizontal gene transfer (HGT) can occur independently of an endosymbiotic event. The publication of the P. tricornutum genome reported that at least 587 P. tricornutum genes appear to be most closely related to bacterial genes, accounting for more than 5% of the P. tricornutum proteome. About half of these are also found in the T. pseudonana genome, attesting their ancient incorporation in the diatom lineage.[94]
Genetic Engineering[]
To understand the biological mechanisms which underlie the great importance of diatoms in geochemical cycles, scientists have used the Phaeodactylum tricornutum and Thalassiosira spp. species as model organisms since the 90's.[98] Few molecular biology tools are currently available to generate mutants or transgenic lines : plasmids containing transgenes are inserted into the cells using the biolistic method[99] or transkingdom bacterial conjugation[100] (with 10−6 and 10−4 yield respectively[99][100]), and other classical transfection methods such as electroporation or use of PEG have been reported to provide results with lower efficiencies.[100]
Transfected plasmids can be either randomly integrated into the diatom's chromosomes or maintained as stable circular episomes (thanks to the CEN6-ARSH4-HIS3 yeast centromeric sequence[100]). The phleomycin/zeocin resistance gene Sh Ble is commonly used as a selection marker,[98][101] and various transgenes have been successfully introduced and expressed in diatoms with stable transmissions through generations,[100][101] or with the possibility to remove it.[101]
Furthermore, these systems now allow the use of the CRISPR-Cas genome edition tool, leading to a fast production of functional knock-out mutants[101][102] and a more accurate comprehension of the diatoms' cellular processes.
Human uses[]
Paleontology[]
Decomposition and decay of diatoms leads to organic and inorganic (in the form of silicates) sediment, the inorganic component of which can lead to a method of analyzing past marine environments by corings of ocean floors or bay muds, since the inorganic matter is embedded in deposition of clays and silts and forms a permanent geological record of such marine strata (see siliceous ooze).
Industrial[]

Diatoms, and their shells (frustules) as diatomite or diatomaceous earth, are important industrial resources used for fine polishing and liquid filtration. The complex structure of their microscopic shells has been proposed as a material for nanotechnology.[103]
Diatomite is considered to be a natural nano material and has many uses and applications such as: production of various ceramic products, construction ceramics, refractory ceramics, special oxide ceramics, for production of humidity control materials, used as filtration material, material in the cement production industry, initial material for production of prolonged-release drug carriers, absorption material in an industrial scale, production of porous ceramics, glass industry, used as catalyst support, as a filler in plastics and paints, purification of industrial waters, pesticide holder, as well as for improving the physical and chemical characteristics of certain soils, and other uses.[104][105][106]
Diatoms are also used to help determine the origin of materials containing them, including seawater.
Forensic[]
The main goal of diatom analysis in forensics is to differentiate a death by submersion from a post-mortem immersion of a body in water. Laboratory tests may reveal the presence of diatoms in the body. Since the silica-based skeletons of diatoms do not readily decay, they can sometimes be detected even in heavily decomposed bodies. As they do not occur naturally in the body, if laboratory tests show diatoms in the corpse that are of the same species found in the water where the body was recovered, then it may be good evidence of drowning as the cause of death. The blend of diatom species found in a corpse may be the same or different from the surrounding water, indicating whether the victim drowned in the same site in which the body was found.[107]
Nanotechnology[]
The deposition of silica by diatoms may also prove to be of utility to nanotechnology.[108] Diatom cells repeatedly and reliably manufacture valves of various shapes and sizes, potentially allowing diatoms to manufacture micro- or nano-scale structures which may be of use in a range of devices, including: optical systems; semiconductor nanolithography; and even vehicles for drug delivery. With an appropriate artificial selection procedure, diatoms that produce valves of particular shapes and sizes might be evolved for cultivation in chemostat cultures to mass-produce nanoscale components.[109] It has also been proposed that diatoms could be used as a component of solar cells by substituting photosensitive titanium dioxide for the silicon dioxide that diatoms normally use to create their cell walls.[110] Diatom biofuel producing solar panels have also been proposed.[111]
History of discovery[]
The first illustrations of diatoms are found in an article from 1703 in Transactions of the Royal Society showing unmistakable drawings of Tabellaria.[112] Although the publication was authored by an unnamed English gentleman, there is recent evidence that he was Charles King of Staffordshire.[112][113] It is only 80 years later that we find the first formally identified diatom, the colonial Bacillaria paxillifera, discovered and described in 1783 by Danish naturalist Otto Friedrich Müller.[112] Like many others after him, he wrongly thought that it was an animal due to its ability to move. Even Charles Darwin saw diatom remains in dust whilst in the Cape Verde Islands, although he was not sure what they were. It was only later that they were identified for him as siliceous polygastrics. The infusoria that Darwin later noted in the face paint of Fueguinos, native inhabitants of Tierra del Fuego in the southern end of South America, were later identified in the same way. During his lifetime, the siliceous polygastrics were clarified as belonging to the Diatomaceae, and Darwin struggled to understand the reasons underpinning their beauty. He exchanged opinions with the noted cryptogamist G. H. K. Thwaites on the topic. In the fourth edition of On the Origin of Species he stated that “Few objects are more beautiful than the minute siliceous cases of the diatomaceae: were these created that they might be examined and admired under the high powers of the microscope”? and reasoned that their exquisite morphologies must have functional underpinnings rather than having been created purely for humans to admire.[114]
References[]
- ^ Dangeard, P. (1933). Traite d'Algologie. Paul Lechvalier and Fils, Paris, [1].
- ^ Dumortier, B.-C. (1822). Commentationes botanicae. Observations botaniques, dédiées à la Société d'Horticulture de Tournay (PDF). Tournay: Imprimerie de Ch. Casterman-Dieu, Rue de pont No. 10. pp. [i], [1]-116, [1, tabl., err.] Archived from the original (PDF) on 6 October 2015 – via Algaebase.
- ^ Rabenhorst, L. Flora europaea algarum aquae dulcis et submarinae (1864–1868). Sectio I. Algas diatomaceas complectens, cum figuris generum omnium xylographice impressis (1864). pp. 1–359. Lipsiae [Leipzig]: Apud Eduardum Kummerum.
- ^ Haeckel, E. (1878). Das Protistenreich.
- ^ Engler, A. & Gilg, E. (1919). Syllabus der Pflanzenfamilien: eine Übersicht über das gesamte Pflanzensystem mit besonderer Berücksichtigung der Medizinal- und Nutzpflanzen, nebst einer Übersicht über die Florenreiche und Florengebiete der Erde zum Gebrauch bei Vorlesungen und Studien über spezielle und medizinisch-pharmazeutische Botanik, 8th ed., Gebrüder Borntraeger Verlag, Berlin, 395 p.
- ^ diá-tom-os "cut in half" (= dichó-tom-os) – diá "through" or "apart" and the root of tém-n-ō "I cut". Alternation between e and o in verb root is ablaut.
- ^ "Definition of DIATOM". www.merriam-webster.com. Archived from the original on 29 July 2018. Retrieved 30 July 2018.
- ^ The Air You're Breathing? A Diatom Made That
- ^ "What are Diatoms?". Diatoms of North America. Archived from the original on 25 January 2020. Retrieved 28 January 2020.
- ^ Treguer, P.; Nelson, D. M.; Van Bennekom, A. J.; Demaster, D. J.; Leynaert, A.; Queguiner, B. (1995). "The Silica Balance in the World Ocean: A Reestimate". Science. 268 (5209): 375–9. Bibcode:1995Sci...268..375T. doi:10.1126/science.268.5209.375. PMID 17746543. S2CID 5672525.
- ^ "King's College London – Lake Megachad". www.kcl.ac.uk. Retrieved 5 May 2018.
- ^ Bristow, C.S.; Hudson-Edwards, K.A.; Chappell, A. (2010). "Fertilizing the Amazon and equatorial Atlantic with West African dust". Geophys. Res. Lett. 37 (14): L14807. Bibcode:2010GeoRL..3714807B. doi:10.1029/2010GL043486.
- ^ Jump up to: a b c d e f g Grethe R. Hasle; Erik E. Syvertsen; Karen A. Steidinger; Karl Tangen (25 January 1996). "Marine Diatoms". In Carmelo R. Tomas (ed.). Identifying Marine Diatoms and Dinoflagellates. Academic Press. pp. 5–385. ISBN 978-0-08-053441-1. Retrieved 13 November 2013.
- ^ "Gas Guzzlers".
- ^ "More on Diatoms". University of California Museum of Paleontology. Archived from the original on 4 October 2012. Retrieved 20 May 2015.
- ^ Nakayama, T.; Kamikawa, R.; Tanifuji, G.; Kashiyama, Y.; Ohkouchi, N.; Archibald, J. M.; Inagaki, Y. (2014). "Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle". Proceedings of the National Academy of Sciences of the United States of America. 111 (31): 11407–11412. Bibcode:2014PNAS..11111407N. doi:10.1073/pnas.1405222111. PMC 4128115. PMID 25049384.
- ^ Pierella Karlusich, Juan José; Pelletier, Eric; Lombard, Fabien; Carsique, Madeline; Dvorak, Etienne; Colin, Sébastien; Picheral, Marc; Cornejo-Castillo, Francisco M.; Acinas, Silvia G.; Pepperkok, Rainer; Karsenti, Eric (6 July 2021). "Global distribution patterns of marine nitrogen-fixers by imaging and molecular methods". Nature Communications. 12 (1): 4160. doi:10.1038/s41467-021-24299-y. ISSN 2041-1723. PMC 8260585. PMID 34230473.
- ^ Wehr, J. D.; Sheath, R. G.; Kociolek, J. P., eds. (2015). Freshwater Algae of North America: Ecology and Classification (2nd ed.). San Diego: Academic Press. ISBN 978-0-12-385876-4.
- ^ Girard, Vincent; Saint Martin, Simona; Buffetaut, Eric; Saint Martin, Jean-Paul; Néraudeau, Didier; Peyrot, Daniel; Roghi, Guido; Ragazzi, Eugenio; Suteethorn, Varavudh (2020). "Thai amber: insights into early diatom history?". BSGF - Earth Sciences Bulletin. 191: 23. doi:10.1051/bsgf/2020028. ISSN 1777-5817.
- ^ Colin, S., Coelho, L.P., Sunagawa, S., Bowler, C., Karsenti, E., Bork, P., Pepperkok, R. and De Vargas, C. (2017) "Quantitative 3D-imaging for cell biology and ecology of environmental microbial eukaryotes". eLife, 6: e26066. doi:10.7554/eLife.26066.002.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ The Inner Space of the Subarctic Pacific Ocean NASA Earth Expeditions, 4 September 2018.
 This article incorporates text from this source, which is in the public domain.
This article incorporates text from this source, which is in the public domain.
- ^ Rousseaux, Cecile S.; Gregg, Watson W. (2015). "Recent decadal trends in global phytoplankton composition". Global Biogeochemical Cycles. 29 (10): 1674–1688. Bibcode:2015GBioC..29.1674R. doi:10.1002/2015GB005139.
- ^ Parker, Andrew R.; Townley, Helen E. (2007). "Biomimetics of photonic nanostructures". Nature Nanotechnology. 2 (6): 347–53. Bibcode:2007NatNa...2..347P. doi:10.1038/nnano.2007.152. PMID 18654305.
- ^ Gordon, Richard; Losic, Dusan; Tiffany, Mary Ann; Nagy, Stephen S.; Sterrenburg, Frithjof A.S. (2009). "The Glass Menagerie: Diatoms for novel applications in nanotechnology". Trends in Biotechnology. 27 (2): 116–27. doi:10.1016/j.tibtech.2008.11.003. PMID 19167770.
- ^ Jump up to: a b c d e Rita A. Horner (2002). A taxonomic guide to some common marine phytoplankton. Biopress. pp. 25–30. ISBN 978-0-948737-65-7. Retrieved 13 November 2013.
- ^ "Glass in Nature". The Corning Museum of Glass. Archived from the original on 7 March 2013. Retrieved 19 February 2013.
- ^ Taylor, J. C., Harding, W. R. and Archibald, C. (2007). An Illustrated Guide to Some Common Diatom Species from South Africa. Gezina: Water Research Commission. ISBN 9781770054844.
- ^ Mishra, M., Arukha, A.P., Bashir, T., Yadav, D. and Prasad, G.B.K.S. (2017) "All new faces of diatoms: potential source of nanomaterials and beyond". Frontiers in microbiology, 8: 1239. doi:10.3389/fmicb.2017.01239.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Jump up to: a b Zhang, D.; Wang, Y.; Cai, J.; Pan, J.; Jiang, X.; Jiang, Y. (2012). "Bio-manufacturing technology based on diatom micro- and nanostructure". Chinese Science Bulletin. 57 (30): 3836–3849. Bibcode:2012ChSBu..57.3836Z. doi:10.1007/s11434-012-5410-x.
- ^ Padisák, Judit; Soróczki-Pintér, Éva; Rezner, Zsuzsanna (2003), Martens, Koen (ed.), "Sinking properties of some phytoplankton shapes and the relation of form resistance to morphological diversity of plankton – an experimental study" (PDF), Aquatic Biodiversity: A Celebratory Volume in Honour of Henri J. Dumont, Developments in Hydrobiology, Springer Netherlands, pp. 243–257, doi:10.1007/978-94-007-1084-9_18, ISBN 9789400710849, archived from the original (PDF) on 23 July 2018, retrieved 4 October 2019
- ^ Anderson, Lars W. J.; Sweeney, Beatrice M. (1 May 1977). "Diel changes in sedimentation characteristics of Ditylum brightwelli: Changes in cellular lipid and effects of respiratory inhibitors and ion-transport modifiers1". Limnology and Oceanography. 22 (3): 539–552. Bibcode:1977LimOc..22..539A. doi:10.4319/lo.1977.22.3.0539. ISSN 1939-5590.
- ^ Poulsen, Nicole C.; Spector, Ilan; Spurck, Timothy P.; Schultz, Thomas F.; Wetherbee, Richard (1 September 1999). "Diatom gliding is the result of an actin-myosin motility system". Cell Motility and the Cytoskeleton. 44 (1): 23–33. doi:10.1002/(SICI)1097-0169(199909)44:1<23::AID-CM2>3.0.CO;2-D. ISSN 1097-0169. PMID 10470016.
- ^ Mann, David G. (February 2010). "raphid diatoms". The Tree of Life Web Project. Retrieved 27 September 2019.
- ^ Jump up to: a b G. Drebes (1 January 1977). "Chapter 9: Sexuality". In Dietrich Werner (ed.). The Biology of Diatoms. Botanical Monographs. 13. University of California Press. pp. 250–283. ISBN 978-0-520-03400-6. Retrieved 14 November 2013.
- ^ Azam, Farooq; Bidle, Kay D. (1999). "Accelerated dissolution of diatom silica by marine bacterial assemblages". Nature. 397 (6719): 508–12. Bibcode:1999Natur.397..508B. doi:10.1038/17351. S2CID 4397909. INIST:1755031.
- ^ Zakharova, Yulia R.; Galachyants, Yuri P.; Kurilkina, Maria I.; Likhoshvay, Alexander V.; Petrova, Darya P.; Shishlyannikov, Sergey M.; Ravin, Nikolai V.; Mardanov, Andrey V.; Beletsky, Alexey V.; Likhoshway, Yelena V. (2013). "The Structure of Microbial Community and Degradation of Diatoms in the Deep Near-Bottom Layer of Lake Baikal". PLOS ONE. 8 (4): e59977. Bibcode:2013PLoSO...859977Z. doi:10.1371/journal.pone.0059977. PMC 3613400. PMID 23560063.
- ^ Jump up to: a b Treguer, P.; Nelson, D. M.; Van Bennekom, A. J.; Demaster, D. J.; Leynaert, A.; Queguiner, B. (1995). "The Silica Balance in the World Ocean: A Reestimate". Science. 268 (5209): 375–9. Bibcode:1995Sci...268..375T. doi:10.1126/science.268.5209.375. PMID 17746543. S2CID 5672525.
- ^ Kilias, Estelle S.; Junges, Leandro; Šupraha, Luka; Leonard, Guy; Metfies, Katja; Richards, Thomas A. (2020). "Chytrid fungi distribution and co-occurrence with diatoms correlate with sea ice melt in the Arctic Ocean". Communications Biology. 3 (1): 183. doi:10.1038/s42003-020-0891-7. PMC 7174370. PMID 32317738. S2CID 216033140.
- ^ Jump up to: a b Egge, J. K.; Aksnes, D. L. (1992). "Silicate as regulating nutrient in phytoplankton competition". Mar. Ecol. Prog. Ser. 83: 281–289. Bibcode:1992MEPS...83..281E. doi:10.3354/meps083281.
- ^ Jump up to: a b c Furnas, Miles J. (1990). "In situ growth rates of marine phytoplankton: Approaches to measurement, community and species growth rates". Journal of Plankton Research. 12 (6): 1117–51. doi:10.1093/plankt/12.6.1117. INIST:5474600.
- ^ Jump up to: a b Yool, Andrew; Tyrrell, Toby (2003). "Role of diatoms in regulating the ocean's silicon cycle". Global Biogeochemical Cycles. 17 (4): n/a. Bibcode:2003GBioC..17.1103Y. CiteSeerX 10.1.1.394.3912. doi:10.1029/2002GB002018.
- ^ Lipps, Jere H. (1970). "Plankton Evolution". Evolution. 24 (1): 1–22. doi:10.2307/2406711. JSTOR 2406711. PMID 28563010.
- ^ Didymo, Aliens Among Us. Archived 7 October 2015 at the Wayback Machine Virtual Exhibit of the Virtual Museum of Canada
- ^ "DEP Reports Didymo Discovered in the West Branch Farmington River. Retrieved on 2014-01-15". Archived from the original on 16 February 2015. Retrieved 27 April 2015.
- ^ "Didymo Stakeholder Update – 31 October 2008". MAF Biosecurity New Zealand www.biosecurity.govt.nz. Archived from the original on 12 February 2013. Retrieved 1 December 2013.
- ^ Jump up to: a b Dugdale, R. C.; Wilkerson, F. P. (1998). "Silicate regulation of new production in the equatorial Pacific upwelling". Nature. 391 (6664): 270–273. Bibcode:1998Natur.391..270D. doi:10.1038/34630. S2CID 4394149.
- ^ Smetacek, V. S. (1985). "Role of sinking in diatom life-history cycles: Ecological, evolutionary and geological significance". Mar. Biol. 84 (3): 239–251. doi:10.1007/BF00392493. S2CID 85054779.
- ^ Raven, J. A. (1983). "The transport and function of silicon in plants". Biol. Rev. 58 (2): 179–207. doi:10.1111/j.1469-185X.1983.tb00385.x. S2CID 86067386.
- ^ Milligan, A. J.; Morel, F. M. M. (2002). "A proton buffering role for silica in diatoms". Science. 297 (5588): 1848–1850. Bibcode:2002Sci...297.1848M. doi:10.1126/science.1074958. PMID 12228711. S2CID 206507070.
- ^ Jump up to: a b Chamberlain, C. J. (1901) Methods in Plant Histology, University of Chicago Press, USA
- ^ Armstrong, E; Rogerson, A; Leftley, Jw (2000). "Utilisation of seaweed carbon by three surface-associated heterotrophic protists, Stereomyxa ramosa, Nitzschia alba and Labyrinthula sp". Aquatic Microbial Ecology. 21: 49–57. doi:10.3354/ame021049. ISSN 0948-3055.
- ^ LEWIN, JOYCE; LEWIN, R. A. (1967). "Culture and Nutrition of Some Apochlorotic Diatoms of the Genus Nitzschia". Microbiology. 46 (3): 361–367. doi:10.1099/00221287-46-3-361. ISSN 1350-0872.
- ^ "Diatoms". Retrieved 13 February 2016.
- ^ Jump up to: a b c Thamatrakoln, K.; Alverson, A.J.; Hildebrand, M. (2006). "Comparative Sequence Analysis of Diatom Silicon Transporters: Toward a Mechanistic Model of Silicon Transport". Journal of Phycology. 42 (4): 822–834. doi:10.1111/j.1529-8817.2006.00233.x. S2CID 86674657.
- ^ Kröger, Nils; Deutzmann, Rainer; Manfred, Sumper (November 1999). "Polycationic Peptides from Diatom Biosilica That Direct Silica Nanosphere Formation". Science. 286 (5442): 1129–1132. doi:10.1126/science.286.5442.1129. PMID 10550045. S2CID 10925689.[permanent dead link]
- ^ Kroger, Nils (2007). Handbook of Biomineralization: Biological Aspects and Structure Formation. Weinheim, Germany: Wiley-VCH Verlag GmbH. pp. chapter 3.
- ^ Allen, Andrew E.; Dupont, Christopher L.; Oborník, Miroslav; Horák, Aleš; Nunes-Nesi, Adriano; McCrow, John P.; Zheng, Hong; Johnson, Daniel A.; Hu, Hanhua; Fernie, Alisdair R.; Bowler, Chris (2011). "Evolution and metabolic significance of the urea cycle in photosynthetic diatoms". Nature. 473 (7346): 203–7. Bibcode:2011Natur.473..203A. doi:10.1038/nature10074. PMID 21562560. S2CID 4350884. Lay summary – Science Daily (12 May 2011).
- ^ Guiry, M.D. (2012). "How many species of algae are there?". Journal of Phycology. 48 (5): 1057–1063. doi:10.1111/j.1529-8817.2012.01222.x. PMID 27011267. S2CID 30911529.
- ^ Frank Eric Round; R. M. Crawford; D. G. Mann (1990). The Diatoms: Biology & Morphology of the Genera. Cambridge University Press. ISBN 978-0-521-36318-1. Retrieved 13 November 2013.[page needed]
- ^ Canter-Lund, H. and Lund, J.W.G. (1995). Freshwater Algae: Their microscopic world explained, Biopress Limited. ISBN 0-948737-25-5.[page needed]
- ^ Mann, David G. (2005). "The species concept in diatoms: Evidence for morphologically distinct, sympatric gamodemes in four epipelic species". Plant Systematics and Evolution. 164 (1/4): 215–37. doi:10.1007/BF00940439. JSTOR 23675282. S2CID 37684109.
- ^ Fourtanier, Elisabeth; Kociolek, J. Patrick (1999). "Catalogue of the Diatom Genera". Diatom Research. 14 (1): 1–190. doi:10.1080/0269249X.1999.9705462.
- ^ The World Register of Marine Species lists 1,356 diatom genus names from all habitats as at July 2020, of which 1,248 are "accepted".
- ^ Queries to the World Register of Marine Species, July 2020, return 299 "fossil only" genus names, of which 285 are "accepted".
- ^ Theriot, Edward C.; Cannone, Jamie J.; Gutell, Robin R.; Alverson, Andrew J. (2009). "The limits of nuclear-encoded SSU rDNA for resolving the diatom phylogeny". European Journal of Phycology. 4 (3): 277–290. doi:10.5091/plecevo.2010.418. PMC 2835975. PMID 20224747.
- ^ Theriot, Edward C.; Ashworth, Matt; Ruck, Elizabeth; Nakov, Teofil; Jansen, Robert K. (2010). "A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research". Plant Ecology and Evolution. 143 (3): 277–290. doi:10.1080/09670260902749159. PMC 2835975. PMID 20224747.
- ^ Parks, Matthew B.; Wickett, Norman J.; Alverson, Andrew J. (2018). "Signal, uncertainty, and conflict in phylogenomic data for a diverse lineage of microbial eukaryotes (Diatoms, Bacillariophyta)". Molecular Biology and Evolution. 35 (1): 80–93. doi:10.1093/molbev/msx268. PMC 5850769. PMID 29040712.
- ^ Medlin, L.K.; Desdevises, Y. (2020). "Review of the phylogenetic reconstruction of the diatoms using molecular tools with an analysis of a seven gene data set using multiple outgroups and morphological data for a total evidence approach" (PDF). Phycologia. in press.
- ^ Adl, Sina M.; Bass, David; Lane, Christopher E.; Lukeš, Julius; Schoch, Conrad L.; Smirnov, Alexey; Agatha, Sabine; Berney, Cedric; Brown, Matthew W. (26 September 2018). "Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes". Journal of Eukaryotic Microbiology. 66 (1): 4–119. doi:10.1111/jeu.12691. PMC 6492006. PMID 30257078.
- ^ Jump up to: a b Kooistra, Wiebe H.C.F.; Medlin, Linda K. (1996). "Evolution of the Diatoms (Bacillariophyta)". Molecular Phylogenetics and Evolution. 6 (3): 391–407. doi:10.1006/mpev.1996.0088. PMID 8975694.
- ^ Schieber, Jürgen; Krinsley, Dave; Riciputi, Lee (2000). "Diagenetic origin of quartz silt in mudstones and implications for silica cycling". Nature. 406 (6799): 981–5. Bibcode:2000Natur.406..981S. doi:10.1038/35023143. PMID 10984049. S2CID 4417951.
- ^ Medlin, L. K.; Kooistra, W. H. C. F.; Gersonde, R.; Sims, P. A.; Wellbrock, U. (1997). "Is the origin of the diatoms related to the end-Permian mass extinction?". Nova Hedwigia. 65 (1–4): 1–11. doi:10.1127/nova.hedwigia/65/1997/1. hdl:10013/epic.12689.
- ^ Raven, J. A.; Waite, A. M. (2004). "The evolution of silicification in diatoms: Inescapable sinking and sinking as escape?". New Phytologist. 162 (1): 45–61. doi:10.1111/j.1469-8137.2004.01022.x. JSTOR 1514475.
- ^ R. Siever; Stephen Henry Schneider; Penelope J. Boston (January 1993). "Silica in the oceans: biological-geological interplay". Scientists on Gaia. MIT Press. pp. 287–295. ISBN 978-0-262-69160-4. Retrieved 14 November 2013.
- ^ Kidder, David L.; Erwin, Douglas H. (2001). "Secular Distribution of Biogenic Silica through the Phanerozoic: Comparison of Silica-Replaced Fossils and Bedded Cherts at the Series Level". The Journal of Geology. 109 (4): 509–22. Bibcode:2001JG....109..509K. doi:10.1086/320794. S2CID 128401816.
- ^ Grenne, Tor; Slack, John F. (2003). "Paleozoic and Mesozoic silica-rich seawater: Evidence from hematitic chert (jasper) deposits". Geology. 31 (4): 319–22. Bibcode:2003Geo....31..319G. doi:10.1130/0091-7613(2003)031<0319:PAMSRS>2.0.CO;2. INIST:14692468.
- ^ Racki, G; Cordey, Fabrice (2000). "Radiolarian palaeoecology and radiolarites: Is the present the key to the past?". Earth-Science Reviews. 52 (1): 83–120. Bibcode:2000ESRv...52...83R. doi:10.1016/S0012-8252(00)00024-6.
- ^ Maldonado, Manuel; Carmona, M. Carmen; Uriz, María J.; Cruzado, Antonio (1999). "Decline in Mesozoic reef-building sponges explained by silicon limitation". Nature. 401 (6755): 785–8. Bibcode:1999Natur.401..785M. doi:10.1038/44560. S2CID 205034177. INIST:1990263.
- ^ Harper, Howard E.; Knoll, Andrew H. (1975). "Silica, diatoms, and Cenozoic radiolarian evolution". Geology. 3 (4): 175–7. Bibcode:1975Geo.....3..175H. doi:10.1130/0091-7613(1975)3<175:SDACRE>2.0.CO;2.
- ^ Falkowski, P. G.; Katz, Miriam E.; Knoll, Andrew H.; Quigg, Antonietta; Raven, John A.; Schofield, Oscar; Taylor, F. J. R. (2004). "The Evolution of Modern Eukaryotic Phytoplankton". Science. 305 (5682): 354–60. Bibcode:2004Sci...305..354F. CiteSeerX 10.1.1.598.7930. doi:10.1126/science.1095964. PMID 15256663. S2CID 451773.
- ^ Kidder, D. L.; Gierlowski-Kordesch, E. H. (2005). "Impact of Grassland Radiation on the Nonmarine Silica Cycle and Miocene Diatomite". PALAIOS. 20 (2): 198–206. Bibcode:2005Palai..20..198K. doi:10.2110/palo.2003.p03-108. JSTOR 27670327. S2CID 140584104.
- ^ Jump up to: a b Lazarus, David; Barron, John; Renaudie, Johan; Diver, Patrick; Türke, Andreas (2014). "Cenozoic Planktonic Marine Diatom Diversity and Correlation to Climate Change". PLOS ONE. 9 (1): e84857. Bibcode:2014PLoSO...984857L. doi:10.1371/journal.pone.0084857. PMC 3898954. PMID 24465441.
- ^ IPCC Core Writing Team, 2007. Climate Change 2007: Synthesis Report. 104.
- ^ Jump up to: a b Scherer, R. P.; Gladenkov, A. Yu.; Barron, J. A. (2007). "Methods and applications of Cenozoic marine diatom biostratigraphy". Paleontological Society Papers. 13: 61–83. doi:10.1017/S1089332600001467.
- ^ Harwood, D. M.; Nikolaev, V. A.; Winter, D. M. (2007). "Cretaceous record of diatom evolution, radiation, and expansion". Paleontological Society Papers. 13: 33–59. doi:10.1017/S1089332600001455.
- ^ Strelnikova, N. I. (1990). "Evolution of diatoms during the Cretaceous and Paleogene periods". In Simola, H. (ed.). Proceedings of the Tenth International Diatom Symposium. Koenigstein: Koeltz Scientific Books. pp. 195–204. ISBN 3-87429-307-6.
- ^ Baldauf, J. G. (1993). "Middle Eocene through early Miocene diatom floral turnover". In Prothero, D.; Berggren, W. H. (eds.). Eocene-Oligocene climatic and biotic evolution. Princeton: Princeton University Press. pp. 310–326. ISBN 0-691-02542-8.
- ^ Barron, J. A. (2003). "Appearance and extinction of planktonic diatoms during the past 18 m.y. in the Pacific and Southern oceans". Diatom Research. 18: 203–224. doi:10.1080/0269249x.2003.9705588. S2CID 84781882.
- ^ Scala, S.; Carels, N; Falciatore, A; Chiusano, M. L.; Bowler, C (2002). "Genome Properties of the Diatom Phaeodactylum tricornutum". Plant Physiology. 129 (3): 993–1002. doi:10.1104/pp.010713. PMC 166495. PMID 12114555.
- ^ Maheswari, U.; Montsant, A; Goll, J; Krishnasamy, S; Rajyashri, K. R.; Patell, V. M.; Bowler, C (2004). "The Diatom EST Database". Nucleic Acids Research. 33 (Database issue): D344–7. doi:10.1093/nar/gki121. PMC 540075. PMID 15608213.
- ^ Montsant, A.; Jabbari, K; Maheswari, U; Bowler, C (2005). "Comparative Genomics of the Pennate Diatom Phaeodactylum tricornutum". Plant Physiology. 137 (2): 500–13. doi:10.1104/pp.104.052829. PMC 1065351. PMID 15665249.
- ^ Maheswari, U.; Mock, T.; Armbrust, E. V.; Bowler, C. (2009). "Update of the Diatom EST Database: A new tool for digital transcriptomics". Nucleic Acids Research. 37 (Database issue): D1001–5. doi:10.1093/nar/gkn905. PMC 2686495. PMID 19029140.
- ^ Jump up to: a b Armbrust, E. V.; Berges, John A.; Bowler, Chris; Green, Beverley R.; Martinez, Diego; Putnam, Nicholas H.; Zhou, Shiguo; Allen, Andrew E.; Apt, Kirk E.; Bechner, Michael; Brzezinski, Mark A.; Chaal, Balbir K.; Chiovitti, Anthony; Davis, Aubrey K.; Demarest, Mark S.; Detter, J. Chris; Glavina, Tijana; Goodstein, David; Hadi, Masood Z.; Hellsten, Uffe; Hildebrand, Mark; Jenkins, Bethany D.; Jurka, Jerzy; Kapitonov, Vladimir V.; Kröger, Nils; Lau, Winnie W. Y.; Lane, Todd W.; Larimer, Frank W.; Lippmeier, J. Casey; et al. (2004). "The Genome of the Diatom Thalassiosira Pseudonana: Ecology, Evolution, and Metabolism". Science. 306 (5693): 79–86. Bibcode:2004Sci...306...79A. CiteSeerX 10.1.1.690.4884. doi:10.1126/science.1101156. PMID 15459382. S2CID 8593895.
- ^ Jump up to: a b c d Bowler, Chris; Allen, Andrew E.; Badger, Jonathan H.; Grimwood, Jane; Jabbari, Kamel; Kuo, Alan; Maheswari, Uma; Martens, Cindy; Maumus, Florian; Otillar, Robert P.; Rayko, Edda; Salamov, Asaf; Vandepoele, Klaas; Beszteri, Bank; Gruber, Ansgar; Heijde, Marc; Katinka, Michael; Mock, Thomas; Valentin, Klaus; Verret, Fréderic; Berges, John A.; Brownlee, Colin; Cadoret, Jean-Paul; Chiovitti, Anthony; Choi, Chang Jae; Coesel, Sacha; De Martino, Alessandra; Detter, J. Chris; Durkin, Colleen; et al. (2008). "The Phaeodactylum genome reveals the evolutionary history of diatom genomes". Nature. 456 (7219): 239–44. Bibcode:2008Natur.456..239B. doi:10.1038/nature07410. PMID 18923393. S2CID 4415177.
- ^ Roy, S. W.; Penny, D. (2007). "A Very High Fraction of Unique Intron Positions in the Intron-Rich Diatom Thalassiosira pseudonana Indicates Widespread Intron Gain". Molecular Biology and Evolution. 24 (7): 1447–57. doi:10.1093/molbev/msm048. PMID 17350938.
- ^ Maumus, Florian; Allen, Andrew E; Mhiri, Corinne; Hu, Hanhua; Jabbari, Kamel; Vardi, Assaf; Grandbastien, Marie-Angèle; Bowler, Chris (2009). "Potential impact of stress activated retrotransposons on genome evolution in a marine diatom". BMC Genomics. 10: 624. doi:10.1186/1471-2164-10-624. PMC 2806351. PMID 20028555.
- ^ Moustafa, A.; Beszteri, B.; Maier, U. G.; Bowler, C.; Valentin, K.; Bhattacharya, D. (2009). "Genomic Footprints of a Cryptic Plastid Endosymbiosis in Diatoms" (PDF). Science. 324 (5935): 1724–6. Bibcode:2009Sci...324.1724M. doi:10.1126/science.1172983. PMID 19556510. S2CID 11408339. Archived from the original (PDF) on 21 April 2014. Retrieved 13 January 2019.
- ^ Jump up to: a b Kroth, Peter G.; Bones, Atle M.; et al. (October 2018). "Genome editing in diatoms : achievements and goals". Plant Cell Reports. 37 (10): 1401–1408. doi:10.1007/s00299-018-2334-1. PMID 30167805. S2CID 52133809.
- ^ Jump up to: a b Falciatore, Angela; Casotti, Raffaella; et al. (May 2015). "Tranformation of Nonselectablporter Genes in Marine Diatomse Re". Marine Biotechnology. 1 (3): 239–251. doi:10.1007/PL00011773. PMID 10383998. S2CID 22267097.
- ^ Jump up to: a b c d e Karas, Bogumil J.; Diner, Rachel E.; et al. (21 April 2015). "Designer diatom episomes delivered by bacterial conjugation". Nature Communications. 6 (1): 6925. Bibcode:2015NatCo...6.6925K. doi:10.1038/ncomms7925. ISSN 2041-1723. PMC 4411287. PMID 25897682.
- ^ Jump up to: a b c d Slattery, Samuel S.; Diamond, Andrew; et al. (16 February 2018). "An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum tricornutum". ACS Synthetic Biology. 7 (2): 328–338. doi:10.1021/acssynbio.7b00191. PMID 29298053.
- ^ Nymark, Marianne; Sharma, Amit Kumar; et al. (July 2016). "A CRISPR/Cas9 system adapted for gene editing in marine algae". Scientific Reports. 6 (1): 24951. Bibcode:2016NatSR...624951N. doi:10.1038/srep24951. PMC 4842962. PMID 27108533.
- ^ Mishra, M; Arukha, AP; Bashir, T; Yadav, D; Gbks, Prasad (2017). "All New Faces of Diatoms: Potential Source of Nanomaterials and Beyond". Front Microbiol. 8: 1239. doi:10.3389/fmicb.2017.01239. PMC 5496942. PMID 28725218.
- ^ Reka, Arianit A.; Pavlovski, Blagoj; Makreski, Petre (October 2017). "New optimized method for low-temperature hydrothermal production of porous ceramics using diatomaceous earth". Ceramics International. 43 (15): 12572–12578. doi:10.1016/j.ceramint.2017.06.132.
- ^ Reka, Arianit; Anovski, Todor; Bogoevski, Slobodan; Pavlovski, Blagoj; Boškovski, Boško (29 December 2014). "Physical-chemical and mineralogical-petrographic examinations of diatomite from deposit near village of Rožden, Republic of Macedonia". Geologica Macedonica. 28 (2): 121–126. ISSN 1857-8586. Archived from the original on 18 April 2020. Retrieved 8 April 2020.
- ^ Reka, Arianit A.; Pavlovski, Blagoj; Ademi, Egzon; Jashari, Ahmed; Boev, Blazo; Boev, Ivan; Makreski, Petre (31 December 2019). "Effect Of Thermal Treatment Of Trepel At Temperature Range 800-1200˚C". Open Chemistry. 17 (1): 1235–1243. doi:10.1515/chem-2019-0132.
- ^ Auer, Antti (1991). "Qualitative Diatom Analysis as a Tool to Diagnose Drowning". The American Journal of Forensic Medicine and Pathology. 12 (3): 213–8. doi:10.1097/00000433-199109000-00009. PMID 1750392. S2CID 38370984.
- ^ Bradbury, J. (2004). "Nature's Nanotechnologists: Unveiling the Secrets of Diatoms". PLOS Biology. 2 (10): 1512–1515. doi:10.1371/journal.pbio.0020306. PMC 521728. PMID 15486572.

- ^ Drum, Ryan W.; Gordon, Richard (2003). "Star Trek replicators and diatom nanotechnology". Trends in Biotechnology. 21 (8): 325–8. doi:10.1016/S0167-7799(03)00169-0. PMID 12902165.
- ^ Johnson, R.C. (9 April 2009). "Diatoms could triple solar cell efficiency". EE Times. Archived from the original on 31 July 2012. Retrieved 13 April 2009.
- ^ Ramachandra, T. V.; Mahapatra, Durga Madhab; b, Karthick; Gordon, Richard (2009). "Milking Diatoms for Sustainable Energy: Biochemical Engineering versus Gasoline-Secreting Diatom Solar Panels". Industrial & Engineering Chemistry Research. 48 (19): 8769–88. doi:10.1021/ie900044j.
- ^ Jump up to: a b c Pierella Karlusich, Juan José; Ibarbalz, Federico M; Bowler, Chris (2020). "Exploration of marine phytoplankton: from their historical appreciation to the omics era". Journal of Plankton Research. 42: 595–612. doi:10.1093/plankt/fbaa049.
- ^ Dolan, John R. (1 August 2019). "Unmasking "The Eldest Son of The Father of Protozoology": Charles King". Protist. 170 (4): 374–384. doi:10.1016/j.protis.2019.07.002. ISSN 1434-4610. PMID 31479910.
- ^ Darwin, Richard (1866). On the Origin of Species by Means of Natural Selection: Or the Preservation of Favoured Races in the Struggle for Life.
External links[]
| Wikispecies has information related to Diatoms. |
| Wikimedia Commons has media related to Diatoms. |
- Diatom EST database, École Normale Supérieure
- Plankton*Net, taxonomic database including images of diatom species
- Life History and Ecology of Diatoms, University of California Museum of Paleontology
- Diatoms: 'Nature's Marbles', Eureka site, University of Bergen
- Diatom life history and ecology, Microfossil Image Recovery and Circulation for Learning and Education (MIRACLE), University College London
- Diatom page Archived 8 October 2009 at the Wayback Machine, Royal Botanic Garden Edinburgh
- Geometry and Pattern in Nature 3: The holes in radiolarian and diatom tests
- Diatom QuickFacts, Monterey Bay Aquarium Research Institute
- Algae image database Academy of Natural Sciences of Philadelphia (ANSP)
- Diatom taxa Academy of Natural Sciences of Philadelphia (ANSP)
- Algae classes
- Diatoms