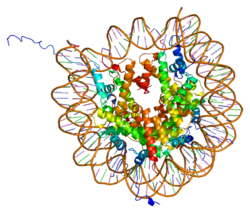
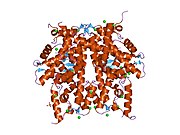
Histone H2A type 1-A is a protein that in humans is encoded by the HIST1H2AA gene .[5] [6]
Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a member of the histone H2A family. Transcripts from this gene contain a palindromic termination element.[6]
References [ ] Further reading [ ]
El Kharroubi A, Piras G, Zensen R, Martin MA (1998). "Transcriptional Activation of the Integrated Chromatin-Associated Human Immunodeficiency Virus Type 1 Promoter" . Mol. Cell. Biol . 18 (5): 2535–44. doi :10.1128/mcb.18.5.2535 . PMC 110633 PMID 9566873 . Deng L, de la Fuente C, Fu P, et al. (2001). "Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones" . Virology . 277 (2): 278–95. doi :10.1006/viro.2000.0593 PMID 11080476 . Deng L, Wang D, de la Fuente C, et al. (2001). "Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA" . Virology . 289 (2): 312–26. doi :10.1006/viro.2001.1129 PMID 11689053 . Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences" . Proc. Natl. Acad. Sci. U.S.A . 99 (26): 16899–903. doi :10.1073/pnas.242603899 PMC 139241 PMID 12477932 . Mungall AJ, Palmer SA, Sims SK, et al. (2003). "The DNA sequence and analysis of human chromosome 6" . Nature . 425 (6960): 805–11. Bibcode :2003Natur.425..805M . doi :10.1038/nature02055 PMID 14574404 . Lusic M, Marcello A, Cereseto A, Giacca M (2004). "Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter" . EMBO J . 22 (24): 6550–61. doi :10.1093/emboj/cdg631 . PMC 291826 PMID 14657027 . Zhang Y, Griffin K, Mondal N, Parvin JD (2004). "Phosphorylation of histone H2A inhibits transcription on chromatin templates" . J. Biol. Chem . 279 (21): 21866–72. doi :10.1074/jbc.M400099200 PMID 15010469 . Aihara H, Nakagawa T, Yasui K, et al. (2004). "Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo" . Genes Dev . 18 (8): 877–88. doi :10.1101/gad.1184604 . PMC 395847 PMID 15078818 . Wang H, Wang L, Erdjument-Bromage H, et al. (2004). "Role of histone H2A ubiquitination in Polycomb silencing". Nature . 431 (7010): 873–8. Bibcode :2004Natur.431..873W . doi :10.1038/nature02985 . PMID 15386022 . S2CID 4344378 . Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)" . Genome Res . 14 (10B): 2121–7. doi :10.1101/gr.2596504 . PMC 528928 PMID 15489334 . Hagiwara T, Hidaka Y, Yamada M (2005). "Deimination of histone H2A and H4 at arginine 3 in HL-60 granulocytes". Biochemistry . 44 (15): 5827–34. doi :10.1021/bi047505c . PMID 15823041 . Cao R, Tsukada Y, Zhang Y (2006). "Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing" . Mol. Cell . 20 (6): 845–54. doi :10.1016/j.molcel.2005.12.002 PMID 16359901 . Bergink S, Salomons FA, Hoogstraten D, et al. (2006). "DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A" . Genes Dev . 20 (10): 1343–52. doi :10.1101/gad.373706 . PMC 1472908 PMID 16702407 .
PDB gallery
1aoi : COMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENT
1eqz : X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION
1hq3 : CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE
1m18 : LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA
1m19 : LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA
1m1a : LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA
1p34 : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3a : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3b : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3f : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3g : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3i : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3k : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3l : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3m : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3o : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1p3p : Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants
1s32 : Molecular Recognition of the Nucleosomal 'Supergroove'
1tzy : Crystal Structure of the Core-Histone Octamer to 1.90 Angstrom Resolution
1zbb : Structure of the 4_601_167 Tetranucleosome
1zla : X-ray Structure of a Kaposi's sarcoma herpesvirus LANA peptide bound to the nucleosomal core
2aro : Crystal Structure Of The Native Histone Octamer To 2.1 Angstrom Resolution, Crystalised In The Presence Of S-Nitrosoglutathione
2cv5 : Crystal structure of human nucleosome core particle
2f8n : 2.9 Angstrom X-ray structure of hybrid macroH2A nucleosomes
2fj7 : Crystal structure of Nucleosome Core Particle Containing a Poly (dA.dT) Sequence Element
2hio : HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN
2nzd : Nucleosome core particle containing 145 bp of DNA