Hyphomicrobiaceae
| Tepidamorphaceae | |
|---|---|
| |
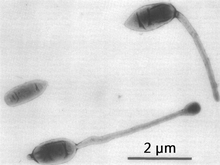
| Three representative cell types of a culture of Hyphomicrobium sp.. Note short processes on two of the three cells, thought to be remnants of the former mother hypha | |
| Scientific classification | |
| Kingdom: | Bacteria
|
| Phylum: | |
| Class: | |
| Order: | |
| Family: | Hyphomicrobiaceae 1950
|
| Genera[3][4] | |
| |
| Synonyms | |
The Hyphomicrobiaceae are a family of bacteria. Among others, they include Rhodomicrobium, a genus of purple bacteria.
Phylogeny[]
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN).[3] The phylogeny is based on whole-genome analysis.[4]
| |||||||||||||||||||||||||||||||||||||
References[]
- ^ Takeuchi M, Katayama T, Yamagishi T, Hanada S, Tamaki H, Kamagata Y, Oshima K, Hattori M, Marumo K, Nedachi M, Maeda H, Suwa Y, Sakata S. (2014). "Methyloceanibacter caenitepidi gen. nov., sp. nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent". Int J Syst Evol Microbiol. 64 (Pt 2): 462–468. doi:10.1099/ijs.0.053397-0. PMID 24096357.CS1 maint: uses authors parameter (link)
- ^ Doronina NV, Poroshina MN, Kaparullina EN, Ezhov VA, Trotsenko YA. (2013). "Methyloligella halotolerans gen. nov., sp. nov. and Methyloligella solikamskensis sp. nov., two non-pigmented halotolerant obligately methylotrophic bacteria isolated from the Ural saline environments". Syst Appl Microbiol. 36 (3): 148–154. doi:10.1016/j.syapm.2012.12.001. PMID 23351489.CS1 maint: uses authors parameter (link)
- ^ Jump up to: a b Euzéby JP, Parte AC. "Hyphomicrobiaceae". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 15 May 2021.CS1 maint: uses authors parameter (link)
- ^ Jump up to: a b Hördt A, García López M, Meier-Kolthoff JP, Schleuning M, Weinhold L-M, Tindall BJ, Gronow A, Kyrpides NC, Woyke T, Göker M. (2020). "Analysis of 1,000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria". Front. Microbiol. 11: 468. doi:10.3389/fmicb.2020.00468.CS1 maint: uses authors parameter (link)
- ^ Jump up to: a b Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. (2014). "Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences". Nat Rev Microbiol. 12 (9): 635–45. doi:10.1038/nrmicro3330. hdl:10261/123763. PMID 25118885. S2CID 21895693.CS1 maint: uses authors parameter (link)
Categories:
- Hyphomicrobiales
- Hyphomicrobiales stubs