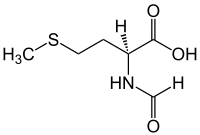
N-Formylmethionine
| |
| Names | |
|---|---|
| IUPAC name
(S)-2-Formylamino-4-methylsulfanylbutanoic acid
| |
| Other names
2-Formylamino-4-methylsulfanyl-butyric acid; Formylmethionine; N-Formyl(methyl)homocysteine
| |
| Identifiers | |
| |
3D model (JSmol)
|
|
| Abbreviations | fMet |
| EC Number |
|
PubChem CID
|
|
| UNII | |
| |
| Properties | |
| C6H11NO3S | |
| Molar mass | 177.22 g/mol |
| Supplementary data page | |
Structure and
properties |
Refractive index (n), Dielectric constant (εr), etc. |
Thermodynamic
data |
Phase behaviour solid–liquid–gas |
Spectral data
|
UV, IR, NMR, MS |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
N-Formylmethionine (fMet,[1] HCO-Met,[2] For-Met[2]) is a derivative of the amino acid methionine in which a formyl group has been added to the amino group. It is specifically used for initiation of protein synthesis from bacterial and organellar genes, and may be removed post-translationally.
fMet plays a crucial part in the protein synthesis of bacteria, mitochondria and chloroplasts. It is not used in cytosolic protein synthesis of eukaryotes, where eukaryotic nuclear genes are translated. It is also not used by Archaea. In the human body, fMet is recognized by the immune system as foreign material, or as an alarm signal released by damaged cells, and stimulates the body to fight against potential infection.
Function in protein synthesis[]
fMet is a starting residue in the synthesis of proteins in bacteria, and, consequently, is located at the N-terminus of the growing polypeptide. fMet is delivered to the ribosome (30S) - mRNA complex by a specialized tRNA (tRNAfMet) which has a 3'-UAC-5' anticodon that is capable of binding with the 5'-AUG-3' start codon located on the mRNA. fMet is thus coded by the same codon as methionine; however, AUG is also the translation initiation codon. When the codon is used for initiation, fMet is used instead of methionine, thereby forming the first amino acid as the peptide chain is synthesized. When the same codon appears later in the mRNA, normal methionine is used. Many organisms use variations of this basic mechanism.
The addition of the formyl group to methionine is catalyzed by the enzyme methionyl-tRNA formyltransferase. This modification is done after methionine has been loaded onto tRNAfMet by aminoacyl-tRNA synthetase.
Methionine itself can be loaded either onto tRNAfMet or tRNAMet. However, transformylase will catalyze the addition of the formyl group to methionine only if methionine has been loaded onto tRNAfMet, not onto tRNAMet.
The N-terminal fMet is removed from majority of proteins, both host and recombinant, by a sequence of two enzymatic reactions. First, peptide deformylase deformylates it, converting the residue back to a normal methionine. Then methionine aminopeptidase (MAP) removes the residue from the chain.[3]
The mitochondria of eukaryotic cells, including those of humans, and the chloroplasts of plant cells also initiate protein synthesis with fMet. Given that mitochondria and chloroplasts have this initial protein synthesis with fMet in common with bacteria, this has been cited as evidence for the endosymbiotic theory.[4]
Relevance to immunology[]
Because fMet is present in proteins made by bacteria but not in those made by eukaryotes (other than in bacterially derived organelles), the immune system might use it to help distinguish self from non-self. Polymorphonuclear cells can bind proteins starting with fMet, and use them to initiate the attraction of circulating blood leukocytes and then stimulate microbicidal activities such as phagocytosis.[5][6][7]
Since fMet is present in proteins made by mitochondria and chloroplasts, more recent theories do not see it as a molecule that the immune system can use to distinguish self from non-self.[8] Instead, fMet-containing oligopeptides and proteins appear to be released by the mitochondria of damaged tissues as well as by damaged bacteria, and can thus qualify as an "alarm" signal, as discussed in the Danger model of immunity. The prototypical fMet-containing oligopeptide is N-Formylmethionine-leucyl-phenylalanine (FMLP) which activates leukocytes and other cell types by binding with these cells' formyl peptide receptor 1 (FPR1) and formyl peptide receptor 2 (FPR2) G protein coupled receptors (see also formyl peptide receptor 3). Acting through these receptors, the fMet-containing oligopeptides and proteins are part of the innate immune system; they function to initiate acute inflammation responses but under other conditions function to inhibit and resolve these responses. fMet-containing oligopeptides and proteins also function in other physiological and pathological responses.
See also[]
References[]
- ^ PubChem. "N-Formyl-DL-methionine". pubchem.ncbi.nlm.nih.gov. Retrieved 2020-10-24.
- ^ a b Nomenclature and Symbolism for Amino Acids and Peptides, 3AA-18 and 3AA-19
- ^ Sherman F, Stewart JW, Tsunasawa S (July 1985). "Methionine or not methionine at the beginning of a protein". BioEssays. 3 (1): 27–31. doi:10.1002/bies.950030108. PMID 3024631. S2CID 33735710.
- ^ Alberts, Bruce (18 November 2014). Molecular biology of the cell (Sixth ed.). New York, NY. p. 800. ISBN 978-0-8153-4432-2. OCLC 887605755.
- ^ Immunology at MCG 1/phagstep
- ^ "The Innate Immune System: Pattern-Recognition Receptors, Antigen-Nonspecific Antimicrobial Body Molecules, and Cytokines". Archived from the original on 2010-07-27.
- ^ Detmers PA, Wright SD, Olsen E, Kimball B, Cohn ZA (September 1987). "Aggregation of complement receptors on human neutrophils in the absence of ligand". The Journal of Cell Biology. 105 (3): 1137–45. doi:10.1083/jcb.105.3.1137. PMC 2114803. PMID 2958480.
- ^ Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ (Mar 4, 2010). "Circulating mitochondrial DAMPs cause inflammatory responses to injury". Nature. 464 (7285): 104–107. Bibcode:2010Natur.464..104Z. doi:10.1038/nature08780. PMC 2843437. PMID 20203610.
External links[]
- N-Formylmethionine at the US National Library of Medicine Medical Subject Headings (MeSH)
- Amino acid derivatives
- Sulfur amino acids
- Formamides