Reprogramming
In biology, reprogramming refers to erasure and remodeling of epigenetic marks, such as DNA methylation, during mammalian development or in cell culture.[1] Such control is also often associated with alternative covalent modifications of histones.
Reprogrammings that are both large scale (10% to 100% of epigenetic marks) and rapid (hours to a few days) occur at three life stages of mammals. Almost 100% of epigenetic marks are reprogrammed in two short periods early in development after fertilization of an ovum by a sperm. In addition, almost 10% of DNA methylations in neurons of the hippocampus can be rapidly altered during formation of a strong fear memory.
After fertilization in mammals, DNA methylation patterns are largely erased and then re-established during early embryonic development. Almost all of the methylations from the parents are erased, first during early embryogenesis, and again in gametogenesis, with demethylation and remethylation occurring each time. Demethylation during early embryogenesis occurs in the preimplantation period. After a sperm fertilizes an ovum to form a zygote, rapid DNA demethylation of the paternal DNA and slower demethylation of the maternal DNA occurs until formation of a morula which has almost no methylation. After the blastocyst is formed, methylation can begin, and with formation of the epiblast a wave of methylation then takes place until the implantation stage of the embryo. Another period of rapid and almost complete demethylation occurs during gametogenesis within the primordial germ cells (PGCs). Other than the PGCs, in the post-implantation stage, methylation patterns in somatic cells are stage- and tissue-specific with changes that presumably define each individual cell type and last stably over a long time.[2]
Embryonic development[]

The mouse sperm genome is 80–90% methylated at its CpG sites in DNA, amounting to about 20 million methylated sites.[3] After fertilization, the paternal chromosome is almost completely demethylated in six hours by an active process, before DNA replication (blue line in Figure). In the mature oocyte, about 40% of its CpG sites are methylated. Demethylation of the maternal chromosome largely takes place by blockage of the methylating enzymes from acting on maternal-origin DNA and by dilution of the methylated maternal DNA during replication (red line in Figure). The morula (at the 16 cell stage), has only a small amount of DNA methylation (black line in Figure). Methylation begins to increase at 3.5 days after fertilization in the blastocyst, and a large wave of methylation then occurs on days 4.5 to 5.5 in the epiblast, going from 12% to 62% methylation, and reaching maximum level after implantation in the uterus.[4] By day seven after fertilization, the newly formed primordial germ cells (PGC) in the implanted embryo segregate from the remaining somatic cells. At this point the PGCs have about the same level of methylation as the somatic cells.
The newly formed primordial germ cells (PGC) in the implanted embryo devolve from the somatic cells. At this point the PGCs have high levels of methylation. These cells migrate from the epiblast toward the gonadal ridge. Now the cells are rapidly proliferating and beginning demethylation in two waves. In the first wave, demethylation is by replicative dilution, but in the second wave demethylation is by an active process. The second wave leads to demethylation of specific loci. At this point the PGC genomes display the lowest levels of DNA methylation of any cells in the entire life cycle [at embryonic day 13.5 (E13.5), see the second figure in this section].[5]

After fertilization some cells of the newly formed embryo migrate to the germinal ridge and will eventually become the germ cells (sperm and oocytes) of the next generation. Due to the phenomenon of genomic imprinting, maternal and paternal genomes are differentially marked and must be properly reprogrammed every time they pass through the germline. Therefore, during the process of gametogenesis the primordial germ cells must have their original biparental DNA methylation patterns erased and re-established based on the sex of the transmitting parent.
After fertilization, the paternal and maternal genomes are demethylated in order to erase their epigenetic signatures and acquire totipotency. There is asymmetry at this point: the male pronucleus undergoes a quick and active demethylation. Meanwhile the female pronucleus is demethylated passively during consecutive cell divisions. The process of DNA demethylation involves base excision repair and likely other DNA-repair-based mechanisms.[6] Despite the global nature of this process, there are certain sequences that avoid it, such as differentially methylated regions (DMRS) associated with imprinted genes, retrotransposons and centromeric heterochromatin. Remethylation is needed again to differentiate the embryo into a complete organism.[7]
In vitro manipulation of pre-implantation embryos has been shown to disrupt methylation patterns at imprinted loci[8] and plays a crucial role in cloned animals.[9]
Learning and Memory[]

Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 h after training, 9.17% of the genes in the rat genomes of hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated.[10] The hippocampus region of the brain is where contextual fear memories are first stored (see figure of the brain, this section), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just 1 day after conditioning, but rats retain a considerable amount of contextual fear when a long delay (28 days) is imposed between the time of conditioning and the time of hippocampectomy.[11]
Molecular stages[]
Three molecular stages are required for reprogramming the DNA methylome. Stage 1: Recruitment. The enzymes needed for reprogramming are recruited to genome sites that require demethylation or methylation. Stage 2: Implementation. The initial enzymatic reactions take place. In the case of methylation, this is a short step that results in the methylation of cytosine to 5-methylcytosine. Stage 3: Base excision DNA repair. The intermediate products of demethylation are catalysed by specific enzymes of the base excision DNA repair pathway that finally restore cystosine in the DNA sequence.
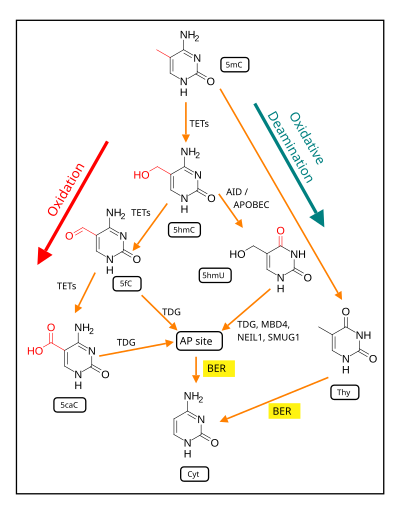
The Figure in this section indicates the central roles of ten-eleven translocation methylcytosine dioxygenases (TETs) in the demethylation of 5-methylcytosine to form cytosine.[13] As reviewed in 2018,[13] 5mC is very often initially oxidized by TET dioxygenases to generate 5-hydroxymethylcytosine (5hmC). In successive steps (see Figure) TET enzymes further hydroxylate 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Thymine-DNA glycosylase (TDG) recognizes the intermediate bases 5fC and 5caC and excises the glycosidic bond resulting in an apyrimidinic site (AP site). In an alternative oxidative deamination pathway, 5hmC can be oxidatively deaminated by APOBEC (AID/APOBEC) deaminases to form 5-hydroxymethyluracil (5hmU) or 5mC can be converted to thymine (Thy). 5hmU can be cleaved by TDG, SMUG1, NEIL1, or MBD4. AP sites and T:G mismatches are then repaired by base excision repair (BER) enzymes to yield cytosine (Cyt).
TET family[]
The isoforms of the TET enzymes include at least two isoforms of TET1, one of TET2 and three isoforms of TET3.[14][15] The full-length canonical TET1 isoform appears virtually restricted to early embryos, embryonic stem cells and primordial germ cells (PGCs). The dominant TET1 isoform in most somatic tissues, at least in the mouse, arises from alternative promoter usage which gives rise to a short transcript and a truncated protein designated TET1s. The isoforms of TET3 are the full length form TET3FL, a short form splice variant TET3s, and a form that occurs in oocytes and neurons designated TET3o. TET3o is created by alternative promoter use and contains an additional first N-terminal exon coding for 11 amino acids. TET3o only occurs in oocytes and neurons and was not expressed in embryonic stem cells or in any other cell type or adult mouse tissue tested. Whereas TET1 expression can barely be detected in oocytes and zygotes, and TET2 is only moderately expressed, the TET3 variant TET3o shows extremely high levels of expression in oocytes and zygotes, but is nearly absent at the 2-cell stage. It is possible that TET3o, high in neurons, oocytes and zygotes at the one cell stage, is the major TET enzyme utilized when very large scale rapid demethylations occur in these cells.
Recruitment of TET to DNA[]
The TET enzymes do not specifically bind to 5-methylcytosine except when recruited. Without recruitment or targeting, TET1 predominantly binds to high CG promoters and CpG islands (CGIs) genome-wide by its CXXC domain that can recognize un-methylated CGIs.[16] TET2 does not have an affinity for 5-methylcytosine in DNA.[17] The CXXC domain of the full-length TET3, which is the predominant form expressed in neurons, binds most strongly to CpGs where the C was converted to 5-carboxycytosine (5caC). However, it also binds to un-methylated CpGs.[15]
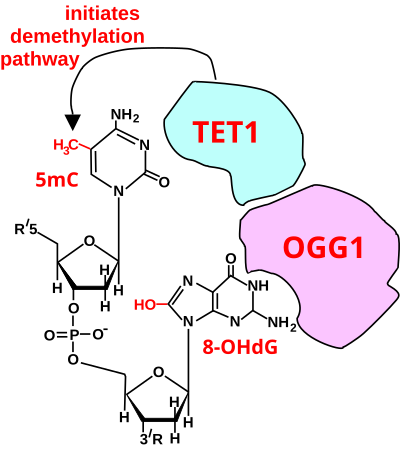
For a TET enzyme to initiate demethylation it must first be recruited to a methylated CpG site in DNA. Two of the proteins shown to recruit a TET enzyme to a methylated cytosine in DNA are OGG1 (see figure Initiation of DNA demthylation)[18] and EGR1.[19]
OGG1[]
Oxoguanine glycosylase (OGG1) catalyses the first step in base excision repair of the oxidatively damaged base 8-OHdG. OGG1 finds 8-OHdG by sliding along the linear DNA at 1,000 base pairs of DNA in 0.1 seconds.[20] OGG1 very rapidly finds 8-OHdG. OGG1 proteins bind to oxidatively damaged DNA with a half maximum time of about 6 seconds.[21] When OGG1 finds 8-OHdG it changes conformation and complexes with 8-OHdG in the binding pocket of OGG1.[22] OGG1 does not immediately act to remove the 8-OHdG. Half maximum removal of 8-OHdG takes about 30 minutes in HeLa cells in vitro,[23] or about 11 minutes in the livers of irradiated mice.[24] DNA oxidation by reactive oxygen species preferentially occurs at a guanine in a methylated CpG site, because of a lowered ionization potential of guanine bases adjacent to 5-methylcytosine.[25] TET1 binds (is recruited to) the OGG1 bound to 8-OHdG (see figure).[18] This likely allows TET1 to demethylate an adjacent methylated cytosine. When human mammary epithelial cells (MCF-10A) were treated with H2O2, 8-OHdG increased in DNA by 3.5-fold and this caused large scale demethylation of 5-methylcytosine to about 20% of its initial level in DNA.[18]
EGR1[]
The gene early growth response protein 1 (EGR1) is an immediate early gene (IEG). The defining characteristic of IEGs is the rapid and transient up-regulation—within minutes—of their mRNA levels independent of protein synthesis.[26] EGR1 can rapidly be induced by neuronal activity.[27] In adulthood, EGR1 is expressed widely throughout the brain, maintaining baseline expression levels in several key areas of the brain including the medial prefrontal cortex, striatum, hippocampus and amygdala.[26] This expression is linked to control of cognition, emotional response, social behavior and sensitivity to reward.[26] EGR1 binds to DNA at sites with the motifs 5′-GCGTGGGCG-3′ and 5'-GCGGGGGCGG-3′ and these motifs occur primarily in promoter regions of genes.[27] The short isoform TET1s is expressed in the brain. EGR1 and TET1s form a complex mediated by the C-terminal regions of both proteins, independently of association with DNA.[27] EGR1 recruits TET1s to genomic regions flanking EGR1 binding sites.[27] In the presence of EGR1, TET1s is capable of locus-specific demethylation and activation of the expression of downstream genes regulated by EGR1.[27]
In cell culture systems[]
Reprogramming can also be induced artificially through the introduction of exogenous factors, usually transcription factors. In this context, it often refers to the creation of induced pluripotent stem cells from mature cells such as adult fibroblasts. This allows the production of stem cells for biomedical research, such as research into stem cell therapies, without the use of embryos. It is carried out by the transfection of stem-cell associated genes into mature cells using viral vectors such as retroviruses.
History[]
The first person to successfully demonstrate reprogramming was John Gurdon, who in 1962 demonstrated that differentiated somatic cells could be reprogrammed back into an embryonic state when he managed to obtain swimming tadpoles following the transfer of differentiated intestinal epithelial cells into enucleated frog eggs.[28] For this achievement he received the 2012 Nobel Prize in Medicine alongside Shinya Yamanaka.[29] Yamanaka was the first to demonstrate (in 2006) that this somatic cell nuclear transfer or oocyte-based reprogramming process (see below), that Gurdon discovered, could be recapitulated (in mice) by defined factors (Oct4, Sox2, Klf4, and c-Myc) to generate induced pluripotent stem cells (iPSCs).[30] Other combinations of genes have also been used.[31]
Variability[]
The properties of cells obtained after reprogramming can vary significantly, in particular among iPSCs.[32] Factors leading to variation in the performance of reprogramming and functional features of end products include genetic background, tissue source, reprogramming factor stoichiometry and stressors related to cell culture.[32]
Somatic cell nuclear transfer[]
An oocyte can reprogram an adult nucleus into an embryonic state after somatic cell nuclear transfer, so that a new organism can be developed from such cell.[33]
Reprogramming is distinct from development of a somatic epitype,[34] as somatic epitypes can potentially be altered after an organism has left the developmental stage of life.[35] During somatic cell nuclear transfer, the oocyte turns off tissue specific genes in the Somatic cell nucleus and turns back on embryonic specific genes.
See also[]
References[]
- ^ Reik W, Dean W, Walter J (August 2001). "Epigenetic reprogramming in mammalian development". Science (Review). 293 (5532): 1089–93. doi:10.1126/science.1063443. PMID 11498579. S2CID 17089710.
- ^ Cedar H, Bergman Y (July 2012). "Programming of DNA methylation patterns". Annual Review of Biochemistry. 81: 97–117. doi:10.1146/annurev-biochem-052610-091920. PMID 22404632.
- ^ "Demethylation in Early Embryonic Development and Memory | IntechOpen".
- ^ Auclair G, Guibert S, Bender A, Weber M (2014). "Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse". Genome Biol. 15 (12): 545. doi:10.1186/s13059-014-0545-5. PMC 4295324. PMID 25476147.
- ^ Zeng Y, Chen T (March 2019). "DNA Methylation Reprogramming during Mammalian Development". Genes (Basel). 10 (4): 257. doi:10.3390/genes10040257. PMC 6523607. PMID 30934924.
- ^ Ladstätter S, Tachibana-Konwalski K (December 2016). "A Surveillance Mechanism Ensures Repair of DNA Lesions during Zygotic Reprogramming". Cell. 167 (7): 1774–1787.e13. doi:10.1016/j.cell.2016.11.009. PMC 5161750. PMID 27916276.
- ^ Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W (January 2013). "Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 368 (1609): 20110330. doi:10.1098/rstb.2011.0330. PMC 3539359. PMID 23166394.
- ^ Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS (September 2003). "Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos". Biology of Reproduction. 69 (3): 902–14. doi:10.1095/biolreprod.103.017293. PMID 12748125.
- ^ Wrenzycki C, Niemann H (December 2003). "Epigenetic reprogramming in early embryonic development: effects of in-vitro production and somatic nuclear transfer". Review. Reproductive Biomedicine Online. 7 (6): 649–56. doi:10.1016/s1472-6483(10)62087-1. PMID 14748963.
- ^ Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017). "Experience-dependent epigenomic reorganization in the hippocampus". Learn. Mem. 24 (7): 278–288. doi:10.1101/lm.045112.117. PMC 5473107. PMID 28620075.
- ^ Kim JJ, Jung MW (2006). "Neural circuits and mechanisms involved in Pavlovian fear conditioning: a critical review". Neurosci Biobehav Rev. 30 (2): 188–202. doi:10.1016/j.neubiorev.2005.06.005. PMC 4342048. PMID 16120461.
- ^ Bayraktar G, Kreutz MR (2018). "The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders". Frontiers in Molecular Neuroscience. 11: 169. doi:10.3389/fnmol.2018.00169. PMC 5975432. PMID 29875631.
- ^ a b Bayraktar G, Kreutz MR (2018). "The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders". Front Mol Neurosci. 11: 169. doi:10.3389/fnmol.2018.00169. PMC 5975432. PMID 29875631.
- ^ Jin SG, Zhang ZM, Dunwell TL, Harter MR, Wu X, Johnson J, Li Z, Liu J, Szabó PE, Lu Q, Xu GL, Song J, Pfeifer GP (January 2016). "Tet3 Reads 5-Carboxylcytosine through Its CXXC Domain and Is a Potential Guardian against Neurodegeneration". Cell Rep. 14 (3): 493–505. doi:10.1016/j.celrep.2015.12.044. PMC 4731272. PMID 26774490.
- ^ a b Melamed P, Yosefzon Y, David C, Tsukerman A, Pnueli L (2018). "Tet Enzymes, Variants, and Differential Effects on Function". Front Cell Dev Biol. 6: 22. doi:10.3389/fcell.2018.00022. PMC 5844914. PMID 29556496.
- ^ Zhang W, Xia W, Wang Q, Towers AJ, Chen J, Gao R, Zhang Y, Yen CA, Lee AY, Li Y, Zhou C, Liu K, Zhang J, Gu TP, Chen X, Chang Z, Leung D, Gao S, Jiang YH, Xie W (December 2016). "Isoform Switch of TET1 Regulates DNA Demethylation and Mouse Development". Mol. Cell. 64 (6): 1062–1073. doi:10.1016/j.molcel.2016.10.030. PMID 27916660.
- ^ Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F (March 2013). "TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS". EMBO J. 32 (5): 645–55. doi:10.1038/emboj.2012.357. PMC 3590984. PMID 23353889.
- ^ a b c d Zhou X, Zhuang Z, Wang W, He L, Wu H, Cao Y, Pan F, Zhao J, Hu Z, Sekhar C, Guo Z (September 2016). "OGG1 is essential in oxidative stress induced DNA demethylation". Cell. Signal. 28 (9): 1163–71. doi:10.1016/j.cellsig.2016.05.021. PMID 27251462.
- ^ Sun Z, Xu X, He J, Murray A, Sun MA, Wei X, Wang X, McCoig E, Xie E, Jiang X, Li L, Zhu J, Chen J, Morozov A, Pickrell AM, Theus MH, Xie H (August 2019). "EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity". Nat Commun. 10 (1): 3892. doi:10.1038/s41467-019-11905-3. PMC 6715719. PMID 31467272.
- ^ Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS (April 2006). "A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA". Proc. Natl. Acad. Sci. U.S.A. 103 (15): 5752–7. doi:10.1073/pnas.0509723103. PMC 1458645. PMID 16585517.
- ^ Abdou I, Poirier GG, Hendzel MJ, Weinfeld M (January 2015). "DNA ligase III acts as a DNA strand break sensor in the cellular orchestration of DNA strand break repair". Nucleic Acids Res. 43 (2): 875–92. doi:10.1093/nar/gku1307. PMC 4333375. PMID 25539916.
- ^ van der Kemp PA, Charbonnier JB, Audebert M, Boiteux S (2004). "Catalytic and DNA-binding properties of the human Ogg1 DNA N-glycosylase/AP lyase: biochemical exploration of H270, Q315 and F319, three amino acids of the 8-oxoguanine-binding pocket". Nucleic Acids Res. 32 (2): 570–8. doi:10.1093/nar/gkh224. PMC 373348. PMID 14752045.
- ^ Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A (September 2004). "In situ analysis of repair processes for oxidative DNA damage in mammalian cells". Proc. Natl. Acad. Sci. U.S.A. 101 (38): 13738–43. doi:10.1073/pnas.0406048101. PMC 518826. PMID 15365186.
- ^ Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A (2001). "A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA". Nucleic Acids Res. 29 (10): 2117–26. doi:10.1093/nar/29.10.2117. PMC 55450. PMID 11353081.
- ^ Ming X, Matter B, Song M, Veliath E, Shanley R, Jones R, Tretyakova N (March 2014). "Mapping structurally defined guanine oxidation products along DNA duplexes: influence of local sequence context and endogenous cytosine methylation". J. Am. Chem. Soc. 136 (11): 4223–35. doi:10.1021/ja411636j. PMC 3985951. PMID 24571128.
- ^ a b c Duclot F, Kabbaj M (2017). "The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders". Front Behav Neurosci. 11: 35. doi:10.3389/fnbeh.2017.00035. PMC 5337695. PMID 28321184.
- ^ a b c d e Sun Z, Xu X, He J, Murray A, Sun MA, Wei X, Wang X, McCoig E, Xie E, Jiang X, Li L, Zhu J, Chen J, Morozov A, Pickrell AM, Theus MH, Xie H (August 2019). "EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity". Nat Commun. 10 (1): 3892. doi:10.1038/s41467-019-11905-3. PMC 6715719. PMID 31467272.
- ^ Gurdon JB (December 1962). "The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles". Journal of Embryology and Experimental Morphology. 10: 622–40. PMID 13951335.
- ^ "The Nobel Prize in Physiology or Medicine – 2012 Press Release". Nobel Media AB. 8 October 2012.
- ^ Takahashi K, Yamanaka S (August 2006). "Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors" (PDF). Cell. 126 (4): 663–76. doi:10.1016/j.cell.2006.07.024. PMID 16904174. S2CID 1565219.
- ^ Baker M (2007-12-06). "Adult cells reprogrammed to pluripotency, without tumors". Nature Reports Stem Cells. doi:10.1038/stemcells.2007.124.
- ^ a b Paull D, Sevilla A, Zhou H, Hahn AK, Kim H, Napolitano C, Tsankov A, Shang L, Krumholz K, Jagadeesan P, Woodard CM, Sun B, Vilboux T, Zimmer M, Forero E, Moroziewicz DN, Martinez H, Malicdan MC, Weiss KA, Vensand LB, Dusenberry CR, Polus H, Sy KT, Kahler DJ, Gahl WA, Solomon SL, Chang S, Meissner A, Eggan K, Noggle SA (September 2015). "Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells". Nature Methods. 12 (9): 885–92. doi:10.1038/nmeth.3507. PMID 26237226. S2CID 9889991.
- ^ Hochedlinger K, Jaenisch R (June 2006). "Nuclear reprogramming and pluripotency". Nature. 441 (7097): 1061–7. Bibcode:2006Natur.441.1061H. doi:10.1038/nature04955. PMID 16810240. S2CID 4304218.
- ^ Lahiri DK, Maloney B (2006). "Genes are not our destiny: the somatic epitype bridges between the genotype and the phenotype". Nature Reviews Neuroscience. 7 (12): 976. doi:10.1038/nrn2022-c1.
- ^ Mathers JC (June 2006). "Nutritional modulation of ageing: genomic and epigenetic approaches". Mechanisms of Ageing and Development. 127 (6): 584–9. doi:10.1016/j.mad.2006.01.018. PMID 16513160. S2CID 9187848.
- DNA
- Epigenetics
- Induced stem cells