Coronavirus membrane protein
| Membrane protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
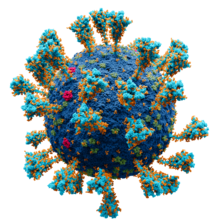 Model of the external structure of the SARS-CoV-2 virion.[1]
● Blue: envelope ● Turquoise: spike glycoprotein (S) ● Red: envelope proteins (E) ● Green: membrane proteins (M) ● Orange: glycans | |||||||||
| Identifiers | |||||||||
| Symbol | CoV_M | ||||||||
| Pfam | PF01635 | ||||||||
| InterPro | IPR002574 | ||||||||
| PROSITE | PS51927 | ||||||||
| |||||||||
The membrane (M) protein (previously called E1, sometimes also matrix protein[2]) is an integral membrane protein that is the most abundant of the four major structural proteins found in coronaviruses.[3][4] The M protein organizes the assembly of coronavirus virions through protein-protein interactions with other M protein molecules as well as with the other three structural proteins, the envelope (E), spike (S), and nucleocapsid (N) proteins.[3][5][6][7]
Structure[]
The M protein is a transmembrane protein with three transmembrane domains and is around 230 amino acid residues long.[7][8] In SARS-CoV-2, the causative agent of Covid-19, the M protein is 222 residues long.[9] Its membrane topology orients the C-terminus toward the cytosolic face of the membrane and thus into the interior of the virion. It has a short N-terminal segment and a larger C-terminal domain. Although the protein sequence is not well conserved across all coronavirus groups, there is a conserved amphipathic region near the C-terminal end of the third transmembrane segment.[7][8]
M functions as a homodimer.[3][4] Studies of the M protein in multiple coronaviruses by cryo-electron microscopy have identified two distinct functional protein conformations, thought to have different roles in forming protein-protein interactions with other structural proteins.[4]
Post-translational modifications[]
M is a glycoprotein whose glycosylation varies according to coronavirus subgroup; N-linked glycosylation is typically found in the alpha and gamma groups while O-linked glycosylation is typically found in the beta group.[7][8] There are some exceptions; for example, in SARS-CoV, a betacoronavirus, the M protein has one N-glycosylation site.[7][5] Glycosylation state does not appear to have a measurable effect on viral growth.[5][8][10] No other post-translational modifications have been described for the M protein.[3]
Expression and localization[]
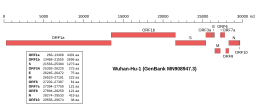 Genomic organisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2, indicating the location of the M gene | |
| NCBI genome ID | 86693 |
|---|---|
| Genome size | 29,903 bases |
| Year of completion | 2020 |
| Genome browser (UCSC) | |
The gene encoding the M protein is located toward the 3' end of the virus's positive-sense RNA genome, along with the genes for the other three structural proteins and various virus-specific accessory proteins.[5][7] M is translated by membrane-bound polysomes[5] to be inserted into the endoplasmic reticulum (ER) and trafficked to the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), the intracellular compartment that gives rise to the coronavirus viral envelope, or to the Golgi apparatus.[7][6][5] The exact localization is dependent on the specific virus protein.[11] Investigations of the subcellular localization of the MERS-CoV M protein found C-terminal sequence signals associated with trafficking to the Golgi.[12]
Function[]

The M protein is the most abundant protein in coronavirus virions.[7][4][3] It is essential for viral replication.[3]
Viral assembly[]
The primary function of the M protein is organizing assembly of new virions.[3] It is involved in establishing viral shape and morphology. Individual M molecules interact with each other to form the viral envelope[6][8][7] and may be able to exclude host cell proteins from the viral membrane.[4] Studies of the SARS-CoV M protein suggest that M-M interactions involve both the N- and C-termini.[5] Coronaviruses are moderately pleomorphic and conformational variations of M appear to be associated with virion size.[4]
M forms protein-protein interactions with all three other major structural proteins.[3][6] M is necessary but not sufficient for viral assembly; M and the E protein expressed together are reportedly sufficient to form virus-like particles,[6] though some reports vary depending on experimental conditions and the specific virus studied.[5][11] In some reports M appears to be capable of inducing membrane curvature,[4] though others report M alone is insufficient for this and E is required.[6] Although the E protein is not necessarily essential, it appears to be required for normal viral morphology and may be responsible for establishing curvature or initiating viral budding.[6] M also appears to have functional roles in the later stages of viral maturation, secretion, and budding.[3]
Incorporation of the spike protein (S) - which is required for assembly of infectious virions - is reported to occur though M interactions and may depend on specific conformations of M.[4][11] The conserved amphipathic region C-terminal to the third transmembrane segment is important for spike interactions.[11] Interactions with M appear to be required for correct subcellular localization of S at the viral budding site.[10] M interacts directly with the nucleocapsid (N) protein without requiring the presence of RNA.[5] This interaction appears to occur primarily through both proteins' C-termini.[3]
Interactions with the immune system[]

The M protein in MERS-CoV, SARS-CoV, and SARS-CoV-2 has been described as an antagonist of interferon response.[3][15]
The M protein is immunogenic and has been reported to be a determinant of humoral immunity.[3] Cytotoxic T cell responses to M have been described.[14] Antibodies to epitopes found in the M protein have been identified in patients recovered from severe acute respiratory syndrome (SARS).[16]
Host cell entry[]
It has been reported that human coronavirus NL63 relies on the M protein as well as the S protein to mediate host cell interactions preceding viral entry. M is thought to bind heparan sulfate proteoglycans exposed on the cell surface.[17]
Evolution and conservation[]
A study of SARS-CoV-2 sequences collected during the Covid-19 pandemic found that missense mutations in the M gene were relatively uncommon and suggested it was under purifying selection.[18] Similar results have been described for broader population genetics analyses over a wider range of related viruses, finding that the sequences of M and several non-structural proteins in the coronavirus genome are most subject to evolutionary constraints.[19]
References[]
- ^ Solodovnikov, Alexey; Arkhipova, Valeria (2021-07-29). "Достоверно красиво: как мы сделали 3D-модель SARS-CoV-2" [Truly beautiful: how we made the SARS-CoV-2 3D model] (in Russian). N+1. Archived from the original on 2021-07-30. Retrieved 30 July 2021.
- ^ Hu, Yongwu; Wen, Jie; Tang, Lin; Zhang, Haijun; Zhang, Xiaowei; Li, Yan; Wang, Jing; Han, Yujun; Li, Guoqing; Shi, Jianping; Tian, Xiangjun; Jiang, Feng; Zhao, Xiaoqian; Wang, Jun; Liu, Siqi; Zeng, Changqing; Wang, Jian; Yang, Huanming (May 2003). "The M Protein of SARS-CoV: Basic Structural and Immunological Properties". Genomics, Proteomics & Bioinformatics. 1 (2): 118–130. doi:10.1016/S1672-0229(03)01016-7. PMC 5172243. PMID 15626342.
- ^ a b c d e f g h i j k l Wong, Nicholas A.; Saier, Milton H. (28 January 2021). "The SARS-Coronavirus Infection Cycle: A Survey of Viral Membrane Proteins, Their Functional Interactions and Pathogenesis". International Journal of Molecular Sciences. 22 (3): 1308. doi:10.3390/ijms22031308. PMC 7865831. PMID 33525632.
- ^ a b c d e f g h Neuman, Benjamin W.; Kiss, Gabriella; Kunding, Andreas H.; Bhella, David; Baksh, M. Fazil; Connelly, Stephen; Droese, Ben; Klaus, Joseph P.; Makino, Shinji; Sawicki, Stanley G.; Siddell, Stuart G.; Stamou, Dimitrios G.; Wilson, Ian A.; Kuhn, Peter; Buchmeier, Michael J. (April 2011). "A structural analysis of M protein in coronavirus assembly and morphology". Journal of Structural Biology. 174 (1): 11–22. doi:10.1016/j.jsb.2010.11.021. PMC 4486061. PMID 21130884.
- ^ a b c d e f g h i Tseng, Ying-Tzu; Wang, Shiu-Mei; Huang, Kuo-Jung; Lee, Amber I-Ru; Chiang, Chien-Cheng; Wang, Chin-Tien (April 2010). "Self-assembly of Severe Acute Respiratory Syndrome Coronavirus Membrane Protein". Journal of Biological Chemistry. 285 (17): 12862–12872. doi:10.1074/jbc.M109.030270. PMC 2857088. PMID 20154085.
- ^ a b c d e f g Schoeman, Dewald; Fielding, Burtram C. (December 2019). "Coronavirus envelope protein: current knowledge". Virology Journal. 16 (1): 69. doi:10.1186/s12985-019-1182-0. PMC 6537279. PMID 31133031.
- ^ a b c d e f g h i Masters, Paul S. (2006). "The Molecular Biology of Coronaviruses". Advances in Virus Research. 66: 193–292. doi:10.1016/S0065-3527(06)66005-3. ISBN 9780120398690. PMC 7112330. PMID 16877062.
- ^ a b c d e J Alsaadi, Entedar A; Jones, Ian M (April 2019). "Membrane binding proteins of coronaviruses". Future Virology. 14 (4): 275–286. doi:10.2217/fvl-2018-0144. PMC 7079996. PMID 32201500.
- ^ Cao, Yipeng; Yang, Rui; Lee, Imshik; Zhang, Wenwen; Sun, Jiana; Wang, Wei; Meng, Xiangfei (June 2021). "Characterization of the SARS‐CoV ‐2 E Protein: Sequence, Structure, Viroporin, and Inhibitors". Protein Science. 30 (6): 1114–1130. doi:10.1002/pro.4075. PMC 8138525. PMID 33813796.
- ^ a b Voß, Daniel; Pfefferle, Susanne; Drosten, Christian; Stevermann, Lea; Traggiai, Elisabetta; Lanzavecchia, Antonio; Becker, Stephan (2009). "Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein". Virology Journal. 6 (1): 79. doi:10.1186/1743-422X-6-79. PMC 2705359. PMID 19534833.
- ^ a b c d Ujike, Makoto; Taguchi, Fumihiro (3 April 2015). "Incorporation of Spike and Membrane Glycoproteins into Coronavirus Virions". Viruses. 7 (4): 1700–1725. doi:10.3390/v7041700. PMC 4411675. PMID 25855243.
- ^ Perrier, Anabelle; Bonnin, Ariane; Desmarets, Lowiese; Danneels, Adeline; Goffard, Anne; Rouillé, Yves; Dubuisson, Jean; Belouzard, Sandrine (September 2019). "The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal". Journal of Biological Chemistry. 294 (39): 14406–14421. doi:10.1074/jbc.RA119.008964. PMC 6768645. PMID 31399512.
- ^ Goodsell, David S.; Voigt, Maria; Zardecki, Christine; Burley, Stephen K. (6 August 2020). "Integrative illustration for coronavirus outreach". PLOS Biology. 18 (8): e3000815. doi:10.1371/journal.pbio.3000815. PMC 7433897. PMID 32760062.
- ^ a b Liu, Jun; Sun, Yeping; Qi, Jianxun; Chu, Fuliang; Wu, Hao; Gao, Feng; Li, Taisheng; Yan, Jinghua; Gao, George F. (15 October 2010). "The Membrane Protein of Severe Acute Respiratory Syndrome Coronavirus Acts as a Dominant Immunogen Revealed by a Clustering Region of Novel Functionally and Structurally Defined Cytotoxic T‐Lymphocyte Epitopes". The Journal of Infectious Diseases. 202 (8): 1171–1180. doi:10.1086/656315. PMC 7537489. PMID 20831383.
- ^ Zheng, Yi; Zhuang, Meng-Wei; Han, Lulu; Zhang, Jing; Nan, Mei-Ling; Zhan, Peng; Kang, Dongwei; Liu, Xinyong; Gao, Chengjiang; Wang, Pei-Hui (December 2020). "Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling". Signal Transduction and Targeted Therapy. 5 (1): 299. doi:10.1038/s41392-020-00438-7. PMC 7768267. PMID 33372174.
- ^ Pang, Hai; Liu, Yinggang; Han, Xueqing; Xu, Yanhui; Jiang, Fuguo; Wu, Donglai; Kong, Xiangang; Bartlam, Mark; Rao, Zihe (1 October 2004). "Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine". Journal of General Virology. 85 (10): 3109–3113. doi:10.1099/vir.0.80111-0. PMID 15448374.
- ^ Naskalska, Antonina; Dabrowska, Agnieszka; Szczepanski, Artur; Milewska, Aleksandra; Jasik, Krzysztof Piotr; Pyrc, Krzysztof (October 2019). "Membrane Protein of Human Coronavirus NL63 Is Responsible for Interaction with the Adhesion Receptor". Journal of Virology. 93 (19): e00355-19. doi:10.1128/JVI.00355-19. PMC 6744225. PMID 31315999.
- ^ Shen, Lishuang; Bard, Jennifer Dien; Triche, Timothy J.; Judkins, Alexander R.; Biegel, Jaclyn A.; Gai, Xiaowu (1 January 2021). "Emerging variants of concern in SARS-CoV-2 membrane protein: a highly conserved target with potential pathological and therapeutic implications". Emerging Microbes & Infections. 10 (1): 885–893. doi:10.1080/22221751.2021.1922097. PMC 8118436. PMID 33896413.
- ^ Cagliani, Rachele; Forni, Diego; Clerici, Mario; Sironi, Manuela (June 2020). "Computational Inference of Selection Underlying the Evolution of the Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2". Journal of Virology. 94 (12): e00411-20. doi:10.1128/JVI.00411-20. PMC 7307108. PMID 32238584.
- Coronavirus proteins
- Viral protein class
- Viral structural proteins
