Diphtheria toxin
| tox diphtheria toxin precursor | |||||||
|---|---|---|---|---|---|---|---|
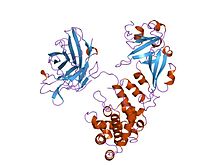 Cartoon representation of the diphtheria toxin protein | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | tox | ||||||
| Entrez | 2650491 | ||||||
| RefSeq (Prot) | NP_938615 | ||||||
| UniProt | P00587 | ||||||
| Other data | |||||||
| EC number | 2.4.2.36 | ||||||
| Chromosome | genome: 0.19 - 0.19 Mb | ||||||
| |||||||
| Diphtheria toxin, C domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Diphtheria_C | ||||||||
| Pfam | PF02763 | ||||||||
| Pfam clan | CL0084 | ||||||||
| InterPro | IPR022406 | ||||||||
| SCOP2 | 1ddt / SCOPe / SUPFAM | ||||||||
| TCDB | 1.C.7 | ||||||||
| |||||||||
| Diphtheria toxin, T domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Diphtheria_T | ||||||||
| Pfam | PF02764 | ||||||||
| InterPro | IPR022405 | ||||||||
| SCOP2 | 1ddt / SCOPe / SUPFAM | ||||||||
| TCDB | 1.C.7 | ||||||||
| |||||||||
| Diphtheria toxin, R domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Diphtheria_R | ||||||||
| Pfam | PF01324 | ||||||||
| InterPro | IPR022404 | ||||||||
| SCOP2 | 1ddt / SCOPe / SUPFAM | ||||||||
| TCDB | 1.C.7 | ||||||||
| |||||||||
Diphtheria toxin is an exotoxin secreted by Corynebacterium, the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage[annotation 1] called corynephage β.[1][2] The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.[3]
Structure[]
Diphtheria toxin is a single polypeptide chain of 535 amino acids consisting of two subunits linked by disulfide bridges, known as an A-B toxin. Binding to the cell surface of the B subunit (the less stable of the two subunits) allows the A subunit (the more stable part of the protein) to penetrate the host cell.[4]
The crystal structure of the diphtheria toxin homodimer has been determined to 2.5 Ångstrom resolution. The structure reveals a Y-shaped molecule consisting of three domains. Fragment A contains the catalytic C domain, and fragment B consists of the T and R domains:[5]
- The amino-terminal catalytic domain, known as the C domain, has an unusual beta+alpha fold.[6] The C domain blocks protein synthesis by transfer of ADP-ribose from NAD to a diphthamide residue of eukaryotic elongation factor 2 (eEF-2).[7][8]
- A central translocation domain, known as the T domain or TM domain, has a multi-helical globin-like fold with two additional helices at the amino terminus but no counterpart to the first globin helix. This domain is thought to unfold in the membrane.[9] A pH-induced conformational change in the T domain triggers insertion into the endosomal membrane and facilitates the transfer of the C domain into the cytoplasm.[7][8]
- A carboxy-terminal receptor-binding domain, known as the R domain, has a beta-sandwich fold consisting of nine strands in two sheets with Greek-key topology; it is a subclass of immunoglobulin-like fold.[6] The R domain binds to a cell surface receptor, permitting the toxin to enter the cell by receptor-mediated endocytosis.[7][8]
Mechanism[]
- Processing
- The leader region is cleaved during secretion.
- Proteolytic nicking separates A and B subunits, which remain joined by disulfide bonds until they reach the cytosol.
- The toxin binds to heparin-binding epidermal growth factor precursor (HB-EGF).
- The complex undergoes endocytosis by the host cell.
- Acidification inside the endosome induces translocation of the A subunit into the cytosol.
- Disulfide bonds are broken.
- The B subunit remains in the endosome as a pore.
- The A subunit ADP-ribosylates host eEF-2. eEF-2 is required for protein synthesis; when it is inactivated, the host cannot make protein and thus dies.
The diphtheria toxin has the same mechanism of action as the enzyme NAD(+)—diphthamide ADP-ribosyltransferase (EC 2.4.2.36). It catalyzes the transfer of NAD+ to a diphthamide residue in eEF-2, inactivating this protein. It does so by ADP-ribosylating the unusual amino acid diphthamide. In this way, it acts as an RNA translational inhibitor. The catalysed reaction is as follows:
- NAD+ + peptide diphthamide nicotinamide + peptide N-(ADP-D-ribosyl)diphthamide.
The exotoxin A of Pseudomonas aeruginosa uses a similar mechanism of action.
Lethal dose and effects[]
Diphtheria toxin is extraordinarily potent.[4] The lethal dose for humans is about 0.1 μg of toxin per kg of body weight. Death occurs through necrosis of the heart and liver.[10] Diphtheria toxin has also been associated with the development of myocarditis. Myocarditis secondary to diphtheria toxin is considered one of the biggest risks to unimmunized children.
History[]
Diphtheria toxin was discovered in 1888 by Émile Roux and Alexandre Yersin. In 1890, Emil Adolf von Behring developed an anti-toxin based on the blood of horses immunized with attenuated bacteria.[11] In 1951, Freeman found that the toxin gene was not encoded on the bacterial chromosome, but by a lysogenic phage (corynephage β)[2] infecting all toxigenic strains.[12][13][14]
Clinical use[]
The drug denileukin diftitox uses diphtheria toxin as an antineoplastic agent.
Resimmune is an immunotoxin that is in clinical trials in cutaneous T cell lymphoma patients. It uses diphtheria toxin (truncated by the cell binding domain) coupled to an antibody to CD3ε (UCHT1).[15]
Research[]
Similar to other A-B toxins, diphtheria toxin is adept at transporting exogenous proteins across mammalian cell membranes, which are usually impermeable to large proteins. This unique ability can be repurposed to deliver therapeutic proteins, instead of the catalytic domain of the toxin.[16][17]
This toxin has also been used in neuroscientific and cancer research to ablate specific populations of cells which express the diphtheria toxin receptor (heparin-binding EGF-like growth factor). Administration of the toxin into the organism which does not naturally express this receptor (e.g. mice) will result in the selective ablation of the cell population which do express it.[18][19]
Annotations[]
- ^ A prophage is a virus that has inserted itself into the genome of the host bacterium.
References[]
- ^ TABLE 1. Bacterial virulence properties altered by bacteriophages from Wagner PL, Waldor MK (August 2002). "Bacteriophage control of bacterial virulence". Infection and Immunity. 70 (8): 3985–93. doi:10.1128/IAI.70.8.3985-3993.2002. PMC 128183. PMID 12117903.
- ^ a b Johnson LP, Tomai MA, Schlievert PM (May 1986). "Bacteriophage Involvement in Group A Streptococcal Pyrogenic Exotoxin A Production". Journal of Bacteriology. 166 (2): 623–7. doi:10.1128/jb.166.2.623-627.1986. PMC 214650. PMID 3009415.
- ^ Bell CE, Eisenberg D (January 1996). "Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide". Biochemistry. 35 (4): 1137–49. doi:10.1021/bi9520848. PMID 8573568.
- ^ a b Murphy JR (1996). "Corynebacterium Diphtheriae: Diphtheria Toxin Production". In Baron S, et al. (eds.). Medical microbiology (4th ed.). Galveston, Texas: Univ. of Texas Medical Branch. ISBN 978-0-9631172-1-2. PMID 21413281.
- ^ Choe S, Bennett MJ, Fujii G, Curmi PM, Kantardjieff KA, Collier RJ, Eisenberg D (May 1992). "The crystal structure of diphtheria toxin". Nature. 357 (6375): 216–22. Bibcode:1992Natur.357..216C. doi:10.1038/357216a0. PMID 1589020. S2CID 4264277.
- ^ a b Bell CE, Eisenberg D (January 1997). "Crystal structure of nucleotide-free diphtheria toxin". Biochemistry. 36 (3): 481–8. CiteSeerX 10.1.1.432.7047. doi:10.1021/bi962214s. PMID 9012663.
- ^ a b c Bennett MJ, Eisenberg D (September 1994). "Refined structure of monomeric diphtheria toxin at 2.3 A resolution". Protein Science. 3 (9): 1464–75. doi:10.1002/pro.5560030912. PMC 2142954. PMID 7833808.
- ^ a b c Bell CE, Eisenberg D (January 1996). "Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide". Biochemistry. 35 (4): 1137–49. doi:10.1021/bi9520848. PMID 8573568.
- ^ Bennett MJ, Choe S, Eisenberg D (September 1994). "Refined structure of dimeric diphtheria toxin at 2.0 A resolution". Protein Science. 3 (9): 1444–63. doi:10.1002/pro.5560030911. PMC 2142933. PMID 7833807.
- ^ Pappenheimer AM (1977). "Diphtheria toxin". Annual Review of Biochemistry. 46 (1): 69–94. doi:10.1146/annurev.bi.46.070177.000441. PMID 20040.
- ^ Enke U (2015). "125 Jahre Diphtherieheilserum: Das Behring'sche Gold" [125 years of diphtheria healing serum: Behring’s gold]. Deutsches Ärzteblatt (in German). 112 (49): A-2088.
- ^ Freeman VJ (June 1951). "Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae". Journal of Bacteriology. 61 (6): 675–88. doi:10.1128/JB.61.6.675-688.1951. PMC 386063. PMID 14850426.
- ^ Freeman VJ, Morse IU (March 1952). "Further observations on the change to virulence of bacteriophage-infected a virulent strains of Corynebacterium diphtheria". Journal of Bacteriology. 63 (3): 407–14. doi:10.1128/JB.63.3.407-414.1952. PMC 169283. PMID 14927573.
- ^ Todar K (2009). "Diphtheria". Todar's Online Textbook of Bacteriology. University of Wisconsin.
- ^ Woo JH, Lee YJ, Neville DM, Frankel AE (2010). "Pharmacology of anti-CD3 diphtheria immunotoxin in CD3 positive T-cell lymphoma trials". Immunotherapy of Cancer. Methods in Molecular Biology. Vol. 651. pp. 157–75. doi:10.1007/978-1-60761-786-0_10. ISBN 978-1-60761-785-3. PMID 20686966.
- ^ Auger A, Park M, Nitschke F, Minassian LM, Beilhartz GL, Minassian BA, Melnyk RA (August 2015). "Efficient Delivery of Structurally Diverse Protein Cargo into Mammalian Cells by a Bacterial Toxin". Molecular Pharmaceutics. 12 (8): 2962–71. doi:10.1021/acs.molpharmaceut.5b00233. PMID 26103531.
- ^ Beilhartz GL, Sugiman-Marangos SN, Melnyk RA (October 2017). "Repurposing bacterial toxins for intracellular delivery of therapeutic proteins". Biochemical Pharmacology. 142: 13–20. doi:10.1016/j.bcp.2017.04.009. PMID 28408344. S2CID 6212879.
- ^ Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, et al. (March 2009). "Selective erasure of a fear memory". Science. 323 (5920): 1492–6. Bibcode:2009Sci...323.1492H. doi:10.1126/science.1164139. PMID 19286560. S2CID 1257448.
- ^ Tammela T, Sage J (2020). "Investigating Tumor Heterogeneity in Mouse Models". Annual Review of Cancer Biology. 4 (1): 99–119. doi:10.1146/annurev-cancerbio-030419-033413. PMC 8218894. PMID 34164589.
External links[]
- Diphtheria+Toxin at the US National Library of Medicine Medical Subject Headings (MeSH)
- How Diphtheria Toxin Works - Animation
- Hepatotoxins
- Protein domains
- Peripheral membrane proteins
- Bacterial toxins
- Diphtheria
- Protein synthesis inhibitors

