trp operon
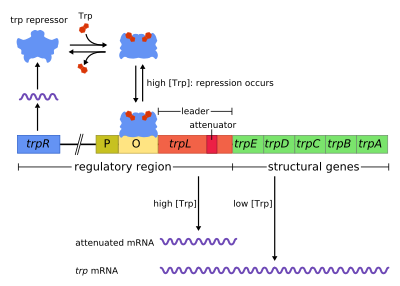
The trp operon is an operon—a group of genes that is used, or transcribed, together—that codes for the components for production of tryptophan. The trp operon is present in many bacteria, but was first characterized in Escherichia coli. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are not expressed. It was an important experimental system for learning about gene regulation, and is commonly used to teach gene regulation.
Trp operon contains five structural genes: trpE, trpD, trpC, trpB, and trpA, which encode enzymatic parts of the pathway. It also contains a repressive regulator gene called trpR. trpR has a promoter where RNA polymerase binds and synthesizes mRNA for a regulatory protein. The protein that is synthesized by trpR then binds to the operator which then causes the transcription to be blocked. In the trp operon, tryptophan binds to the repressor protein effectively blocking gene transcription. In this situation, repression is that of RNA polymerase transcribing the genes in the operon. Also unlike the lac operon, the trp operon contains a leader peptide and an attenuator sequence which allows for graded regulation.[1]
It is an example of repressible negative regulation of gene expression. Within the operon's regulatory sequence, the operator is bound to the repressor protein in the presence of tryptophan (thereby preventing transcription) and is liberated in tryptophan's absence (thereby allowing transcription).
Genes[]
Trp operon contains five structural genes. Their roles are:
- TrpE (P00895): Anthranilate synthase produces anthranilate.
- TrpD (P00904): Cooperates with TrpE.
- TrpC (P00909): Phosphoribosylanthranilate isomerase domain first turns N-(5-phospho-β-D-ribosyl)anthranilate into 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate. The Indole-3-glycerol-phosphate synthase on the same protein then turns the product into (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate.
- TrpA (P0A877), TrpB (P0A879): two subunits of tryptophan synthetase. Combines TrpC's product with serine to produce tryptophan.
Repression[]

The operon operates by a negative repressible feedback mechanism. The repressor for the trp operon is produced upstream by the trpR gene, which is constitutively expressed at a low level. Synthesized trpR monomers associate into dimers. When tryptophan is present, these tryptophan repressor dimers bind to tryptophan, causing a change in the repressor conformation, allowing the repressor to bind to the operator. This prevents RNA polymerase from binding to and transcribing the operon, so tryptophan is not produced from its precursor. When tryptophan is not present, the repressor is in its inactive conformation and cannot bind the operator region, so transcription is not inhibited by the repressor.
Attenuation[]

Attenuation is a second mechanism of negative feedback in the trp operon. The repression system targets the intracellular trp concentration whereas the attenuation responds to the concentration of charged tRNAtrp.[2] Thus, the trpR repressor decreases gene expression by altering the initiation of transcription, while attenuation does so by altering the process of transcription that's already in progress.[2] While the TrpR repressor decreases transcription by a factor of 70, attenuation can further decrease it by a factor of 10, thus allowing accumulated repression of about 700-fold.[3] Attenuation is made possible by the fact that in prokaryotes (which have no nucleus), the ribosomes begin translating the mRNA while RNA polymerase is still transcribing the DNA sequence. This allows the process of translation to affect transcription of the operon directly.
At the beginning of the transcribed genes of the trp operon is a sequence of at least 130 nucleotides termed the leader transcript (trpL; P0AD92).[4] Lee and Yanofsky (1977) found that the attenuation efficiency is correlated with the stability of a secondary structure embedded in trpL,[5] and the 2 constituent hairpins of the terminator structure were later elucidated by Oxender et al. (1979).[6] This transcript includes four short sequences designated 1–4, each of which is partially complementary to the next one. Thus, three distinct secondary structures (hairpins) can form: 1–2, 2–3 or 3–4. The hybridization of sequences 1 and 2 to form the 1–2 structure is rare because the RNA polymerase waits for a ribosome to attach before continuing transcription past sequence 1, however if the 1–2 hairpin were to form it would prevent the formation of the 2–3 structure (but not 3–4). The formation of a hairpin loop between sequences 2–3 prevents the formation of hairpin loops between both 1–2 and 3–4. The 3–4 structure is a transcription termination sequence (abundant in G/C and immediately followed by several uracil residues), once it forms RNA polymerase will disassociate from the DNA and transcription of the structural genes of the operon can not occur (see below for a more detailed explanation). The functional importance of the 2nd hairpin for the transcriptional termination is illustrated by the reduced transcription termination frequency observed in experiments destabilizing the central G+C pairing of this hairpin.[5][7][8][9]
Part of the leader transcript codes for a short polypeptide of 14 amino acids, termed the leader peptide. This peptide contains two adjacent tryptophan residues, which is unusual, since tryptophan is a fairly uncommon amino acid (about one in a hundred residues in a typical E. coli protein is tryptophan). The strand 1 in trpL encompasses the region encoding the trailing residues of the leader peptide: Trp, Trp, Arg, Thr, Ser;[2] conservation is observed in these 5 codons whereas mutating the upstream codons do not alter the operon expression.[2][10][11][12] If the ribosome attempts to translate this peptide while tryptophan levels in the cell are low, it will stall at either of the two trp codons. While it is stalled, the ribosome physically shields sequence 1 of the transcript, preventing the formation of the 1–2 secondary structure. Sequence 2 is then free to hybridize with sequence 3 to form the 2–3 structure, which then prevents the formation of the 3–4 termination hairpin, which is why the 2–3 structure is called an anti-termination hairpin. In the presence of the 2–3 structure, RNA polymerase is free to continue transcribing the operon. Mutational analysis and studies involving complementary oligonucleotides demonstrate that the stability of the 2–3 structure corresponds to the operon expression level.[10][13][14][15] If tryptophan levels in the cell are high, the ribosome will translate the entire leader peptide without interruption and will only stall during translation termination at the stop codon. At this point the ribosome physically shields both sequences 1 and 2. Sequences 3 and 4 are thus free to form the 3–4 structure which terminates transcription. This terminator structure forms when no ribosome stalls in the vicinity of the Trp tandem (i.e. Trp or Arg codon): either the leader peptide is not translated or the translation proceeds smoothly along the strand 1 with abundant charged tRNAtrp.[2][10] More over, the ribosome is proposed to only block about 10 nts downstream, thus ribosome stalling in either the upstream Gly or further downstream Thr do not seem to affect the formation of the termination hairpin.[2][10] The end result is that the operon will be transcribed only when tryptophan is unavailable for the ribosome, while the trpL transcript is constitutively expressed.
This attenuation mechanism is experimentally supported. First, the translation of the leader peptide and ribosomal stalling are directly evidenced to be necessary for inhibiting the transcription termination.[13] Moreover, mutational analysis destabilizing or disrupting the base-pairing of the antiterminator hairpin results in increased termination of several folds; consistent with the attenuation model, this mutation fails to relieve attenuation even with starved Trp.[10][13] In contrast, complementary oligonucleotides targeting strand 1 increases the operon expression by promoting the antiterminator formation.[10][14] Furthermore, in histidine operon, compensatory mutation shows that the pairing ability of strands 2–3 matters more than their primary sequence in inhibiting attenuation.[10][15]
In attenuation, where the translating ribosome is stalled determines whether the termination hairpin will be formed.[10] In order for the transcribing polymerase to concomitantly capture the alternative structure, the time scale of the structural modulation must be comparable to that of the transcription.[2] To ensure that the ribosome binds and begins translation of the leader transcript immediately following its synthesis, a pause site exists in the trpL sequence. Upon reaching this site, RNA polymerase pauses transcription and apparently waits for translation to begin. This mechanism allows for synchronization of transcription and translation, a key element in attenuation.
A similar attenuation mechanism regulates the synthesis of histidine, phenylalanine and threonine.
Regulation of trp operon in Bacillus subtilis[]
Trp operon genes are arranged in the same order in E. coli and Bacillus subtilis.[16] Regulation of trp operons in both organisms depends on the amount of trp present in the cell. However, the primary regulation of tryptophan biosynthesis in B. subtilis is via attenuation, rather than repression, of transcription.[17] In B. subtilis, tryptophan binds to the eleven-subunit tryptophan-activated RNA-binding attenuation protein (TRAP), which activates TRAP's ability to bind to the trp leader RNA.[18][19] Binding of trp-activated TRAP to leader RNA results in the formation of a terminator structure that causes transcription termination.[17]
References[]
- ^ Klug WS, Cummings MR, Spencer C (2006). Concepts of Genetics (8th ed.). New Jersey: Pearson Education Inc. pp. 394–402. ISBN 978-0-13-191833-7.
- ^ a b c d e f g Yanofsky C (February 1981). "Attenuation in the control of expression of bacterial operons". Nature. 289 (5800): 751–8. Bibcode:1981Natur.289..751Y. doi:10.1038/289751a0. PMID 7007895. S2CID 4364204.
- ^ Lehninger AL, Nelson DL, Cox MM (2008). Principles of Biochemistry (5th ed.). New York, NY: W.H. Freeman and Company. p. 1128. ISBN 978-0-7167-7108-1.
- ^ Bertrand K, Squires C, Yanofsky C (May 1976). "Transcription termination in vivo in the leader region of the tryptophan operon of Escherichia coli". Journal of Molecular Biology. 103 (2): 319–37. doi:10.1016/0022-2836(76)90315-6. PMID 781269.
- ^ a b Lee F, Yanofsky C (October 1977). "Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination". Proceedings of the National Academy of Sciences of the United States of America. 74 (10): 4365–9. Bibcode:1977PNAS...74.4365L. doi:10.1073/pnas.74.10.4365. PMC 431942. PMID 337297.
- ^ Oxender DL, Zurawski G, Yanofsky C (November 1979). "Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region". Proceedings of the National Academy of Sciences of the United States of America. 76 (11): 5524–8. Bibcode:1979PNAS...76.5524O. doi:10.1073/pnas.76.11.5524. PMC 411681. PMID 118451.
- ^ Stroynowski I, Yanofsky C (July 1982). "Transcript secondary structures regulate transcription termination at the attenuator of S. marcescens tryptophan operon". Nature. 298 (5869): 34–8. Bibcode:1982Natur.298...34S. doi:10.1038/298034a0. PMID 7045685. S2CID 4347442.
- ^ Zurawski G, Yanofsky C (September 1980). "Escherichia coli tryptophan operon leader mutations, which relieve transcription termination, are cis-dominant to trp leader mutations, which increase transcription termination". Journal of Molecular Biology. 142 (1): 123–9. doi:10.1016/0022-2836(80)90210-7. PMID 6159477.
- ^ Stauffer GV, Zurawski G, Yanofsky C (October 1978). "Single base-pair alterations in the Escherichia coli trp operon leader region that relieve transcription termination at the trp attenuator". Proceedings of the National Academy of Sciences of the United States of America. 75 (10): 4833–7. Bibcode:1978PNAS...75.4833S. doi:10.1073/pnas.75.10.4833. PMC 336215. PMID 368800.
- ^ a b c d e f g h Kolter R, Yanofsky C (1982). "Attenuation in amino acid biosynthetic operons". Annual Review of Genetics. 16: 113–34. doi:10.1146/annurev.ge.16.120182.000553. PMID 6186194.
- ^ Lee F, Bertrand K, Bennett G, Yanofsky C (May 1978). "Comparison of the nucleotide sequences of the initial transcribed regions of the tryptophan operons of Escherichia coli and Salmonella typhimurium". Journal of Molecular Biology. 121 (2): 193–217. doi:10.1016/s0022-2836(78)80005-9. PMID 351195.
- ^ Miozzari G, Yanofsky C (November 1978). "Naturally occurring promoter down mutation: nucleotide sequence of the trp promoter/operator/leader region of Shigella dysenteriae 16". Proceedings of the National Academy of Sciences of the United States of America. 75 (11): 5580–4. Bibcode:1978PNAS...75.5580M. doi:10.1073/pnas.75.11.5580. PMC 393010. PMID 364484.
- ^ a b c Zurawski G, Elseviers D, Stauffer GV, Yanofsky C (December 1978). "Translational control of transcription termination at the attenuator of the Escherichia coli tryptophan operon". Proceedings of the National Academy of Sciences of the United States of America. 75 (12): 5988–92. Bibcode:1978PNAS...75.5988Z. doi:10.1073/pnas.75.12.5988. PMC 393102. PMID 366606.
- ^ a b Winkler ME, Mullis K, Barnett J, Stroynowski I, Yanofsky C (April 1982). "Transcription termination at the tryptophan operon attenuator is decreased in vitro by an oligomer complementary to a segment of the leader transcript". Proceedings of the National Academy of Sciences of the United States of America. 79 (7): 2181–5. Bibcode:1982PNAS...79.2181W. doi:10.1073/pnas.79.7.2181. PMC 346154. PMID 6179092.
- ^ a b Johnston HM, Roth JR (February 1981). "DNA sequence changes of mutations altering attenuation control of the histidine operon of Salmonella typhimurium". Journal of Molecular Biology. 145 (4): 735–56. doi:10.1016/0022-2836(81)90312-0. PMID 6167727.
- ^ Merino E, Jensen RA, Yanofsky C (April 2008). "Evolution of bacterial trp operons and their regulation". Current Opinion in Microbiology. 11 (2): 78–86. doi:10.1016/j.mib.2008.02.005. PMC 2387123. PMID 18374625.
- ^ a b Gollnick P, Babitzke P, Antson A, Yanofsky C (2005-11-14). "Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis". Annual Review of Genetics. 39 (1): 47–68. doi:10.1146/annurev.genet.39.073003.093745. PMID 16285852.
- ^ Elliott MB, Gottlieb PA, Gollnick P (January 2001). "The mechanism of RNA binding to TRAP: initiation and cooperative interactions". RNA. 7 (1): 85–93. doi:10.1017/S135583820100173X. PMC 1370072. PMID 11214184.
- ^ Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, et al. (April 1995). "The structure of trp RNA-binding attenuation protein". Nature. 374 (6524): 693–700. Bibcode:1995Natur.374..693A. doi:10.1038/374693a0. PMID 7715723. S2CID 4340136.
Further reading[]
- Morse DE, Mosteller RD, Yanofsky C (1969). "Dynamics of synthesis, translation, and degradation of trp operon messenger RNA in E. coli". Cold Spring Harbor Symposia on Quantitative Biology. 34: 725–40. doi:10.1101/sqb.1969.034.01.082. PMID 4909527.
- Yanofsky C (February 1981). "Attenuation in the control of expression of bacterial operons". Nature. 289 (5800): 751–8. Bibcode:1981Natur.289..751Y. doi:10.1038/289751a0. PMID 7007895. S2CID 4364204.
External links[]
- Gene expression
- Operons