H5N1 genetic structure
hideThis article has multiple issues. Please help or discuss these issues on the talk page. (Learn how and when to remove these template messages)
|
 |
H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA.
H5N1 is an Influenza A virus subtype. Experts believe it might mutate into a form that transmits easily from person to person. If such a mutation occurs, it might remain an H5N1 subtype or could shift subtypes as did H2N2 when it evolved into the Hong Kong Flu strain of H3N2.
H5N1 has mutated[1] through antigenic drift into dozens of highly pathogenic varieties, but all currently belonging to genotype Z of avian influenza virus H5N1. Genotype Z emerged through reassortment in 2002 from earlier highly pathogenic genotypes of H5N1 that first appeared in China in 1996 in birds and in Hong Kong in 1997 in humans.[2] The "H5N1 viruses from human infections and the closely related avian viruses isolated in 2004 and 2005 belong to a single genotype, often referred to as genotype Z."[1]
This infection of humans coincided with an epizootic (an epidemic in nonhumans) of H5N1 influenza in Hong Kong's poultry population. This panzootic (a disease affecting animals of many species especially over a wide area) outbreak was stopped by the killing of the entire domestic poultry population within the territory. The name H5N1 refers to the subtypes of surface antigens present on the virus: hemagglutinin type 5 and neuraminidase type 1.
Genotype Z of H5N1 is now the dominant genotype of H5N1. Genotype Z is endemic in birds in southeast Asia and represents a long term pandemic threat.
Influenza A viruses have 11 genes on eight separate RNA molecules [1]:
- PB2 (polymerase basic 2)
- PB1 (polymerase basic 1)
- PB1-F2 (alternate open reading frame near the 5' end of the PB1 gene)
- PA (polymerase acidic)
- HA (hemagglutinin)
- NP (nucleoprotein)
- NA (neuraminidase)
- M1 and M2 (matrix)
- NS1 (non-structural)
- NEP/NS2 (nuclear export of vRNPs)
Two of the most important RNA molecules are HA and PB1. HA creates a surface antigen that is especially important in transmissibility. PB1 creates a viral polymerase molecule that is especially important in virulence.
The HA RNA molecule contains the HA gene, which codes for hemagglutinin, which is an antigenic glycoprotein found on the surface of the influenza viruses and is responsible for binding the virus to the cell that is being infected. Hemagglutinin forms spikes at the surface of flu viruses that function to attach viruses to cells. This attachment is required for efficient transfer of flu virus genes into cells, a process that can be blocked by antibodies that bind to the hemagglutinin proteins.
One genetic factor in distinguishing between human flu viruses and avian flu viruses is that avian influenza HA bind alpha 2-3 sialic acid receptors while human influenza HA bind alpha 2-6 sialic acid receptors. Swine influenza viruses have the ability to bind both types of sialic acid receptors. Humans have avian-type receptors at very low densities and chickens have human-type receptors at very low densities. Some isolates taken from H5N1-infected human have been observed to have HA mutations at positions 182, 192, 223, 226, or 228 and these mutations have been shown to influence the selective binding of the virus to those previously mentioned sialic acid avian and/or human cell surface receptors. These are the types of mutations that can change a bird flu virus into a flu pandemic virus.
A 2008 virulence study that mated in a laboratory an avian flu H5N1 virus that circulated in Thailand in 2004 and a human flu H3N2 virus recovered in Wyoming in 2003 produced 63 viruses representing various potential combinations of human and avian influenza A virus genes. One in five were lethal to mice at low doses. The virus that most closely matched H5N1 for virulence was one with the hemagglutinin (HA), the neuraminidase (NA) and the PB1 avian flu virus RNA molecules with their genes combined with the remaining five RNA molecules (PB2, PA, NP, M, and NS) with their genes from the human flu virus. Both the viruses from the 1957 pandemic and 1968 pandemic carried an avian flu virus PB1 gene. The authors suggest that picking up an avian flu virus PB1 gene may be a critical step in a potential flu pandemic virus arising through reassortment."[3]
PB1 codes for the PB1 protein and the PB1-F2 protein. The PB1 protein is a critical component of the viral polymerase. The PB1-F2 protein is encoded by an alternative open reading frame of the PB1 RNA segment and "interacts with 2 components of the mitochondrial permeability transition pore complex, ANT3 and VDCA1, [sensitizing] cells to apoptosis. [...] PB1-F2 likely contributes to viral pathogenicity and might have an important role in determining the severity of pandemic influenza."[4] This was discovered by Chen et al. and reported in Nature.[5] "After comparing viruses from the Hong Kong 1997 H5N1 outbreak, one amino acid change (N66S) was found in the PB1-F2 sequence at position 66 that correlated with pathogenicity. This same amino acid change (N66S) was also found in the PB1-F2 protein of the 1918 pandemic A/Brevig Mission/18 virus."[6]
Terminology[]
The Orthomyxovirus family consists of 5 genera: Influenzavirus A, Influenzavirus B, Influenzavirus C, Isavirus, and Thogotovirus.
The "RNA viruses" include the "negative-sense ssRNA viruses" which include the Family "Orthomyxoviridae" which contains five genera, classified by variations in nucleoprotein (NP and M) antigens. One of these is the Genus "Influenzavirus A" which consists of a single species called "Influenza A virus"; one of its subtypes is H5N1.
H5N1 (like the other avian flu viruses) has strains called "highly pathogenic" (HP) and "low-pathogenic" (LP). Avian influenza viruses that cause HPAI are highly virulent, and mortality rates in infected flocks often approach 100%. LPAI viruses are generally of lower virulence, but these viruses can serve as progenitors to HPAI viruses. The current strain of H5N1 responsible for die-offs of domestic birds in Asia is an HPAI strain; other strains of H5N1 occurring elsewhere in the world are less virulent and, therefore, are classified as LPAI strains. All HPAI strains identified to date have involved H5 and H7 subtypes. The distinction concerns pathogenicity in poultry, not humans. Normally a highly pathogenic avian virus is not highly pathogenic to either humans or non-poultry birds. This current strain of H5N1 is unusual in being deadly to so many species.
Both "influenza" (meaning flu) and "A" (meaning species type A) can be used as adjectives of the noun "virus" resulting in the noun phrase "influenza A virus"; which when capitalized is the proper noun Influenza A virus which is the name of the species the noun phrase also refers to.
Context[]
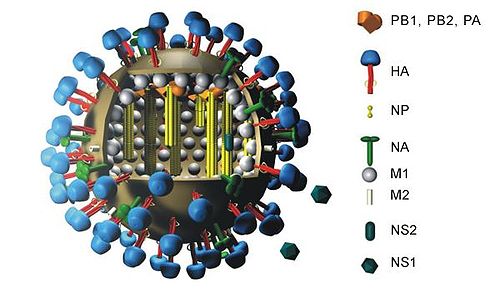
- Virus
A virus is one type of microscopic parasite that infects cells in biological organisms.
- Orthomyxoviridae
The Orthomyxoviridae are a family of RNA viruses which infect vertebrates. It includes those viruses which cause influenza. Viruses of this family contain 7 to 8 segments of linear negative-sense single-stranded RNA.
- Influenza virus
"Influenza virus" refers to a subset of Orthomyxoviridae that create influenza. This taxonomic category is not based on phylogenetics.
- Influenza A virus
Influenza A viruses have 10 genes on eight separate RNA molecules, which, for the reasons mentioned above, are named PB2, PB1, PA, HA, NP, NA, M, and NS. HA, NA, and M specify the structure of proteins that are most medically relevant as targets for antiviral drugs and antibodies. (An eleventh recently discovered gene called PB1-F2 sometimes creates a protein but is absent from some influenza virus isolates.[5]) This segmentation of the influenza genome facilitates genetic recombination by segment reassortment in hosts who are infected with two different influenza viruses at the same time.[1] Influenza A virus is the only species in the Influenzavirus A genus of the family Orthomyxoviridae and are negative sense, single-stranded, segmented RNA viruses.
"The influenza virus RNA polymerase is a multifunctional complex composed of the three viral proteins PB1, PB2 and PA, which, together with the viral nucleoprotein NP, form the minimum complement required for viral mRNA synthesis and replication."[7]
Surface encoding gene segments[]
- Surface antigen encoding gene segments (RNA molecule): (HA, NA)
- HA codes for hemagglutinin, which is an antigenic glycoprotein found on the surface of the influenza viruses and is responsible for binding the virus to the cell that is being infected. Hemagglutinin forms spikes at the surface of flu viruses that function to attach viruses to cells. This attachment is required for efficient transfer of flu virus genes into cells, a process that can be blocked by antibodies that bind to the hemagglutinin proteins. One genetic factor in distinguishing between human flu viruses and avian flu viruses is that "avian influenza HA bind alpha 2-3 sialic acid receptors while human influenza HA bind alpha 2-6 sialic acid receptors. Swine influenza viruses have the ability to bind both types of sialic acid receptors."[8] A mutation found in Turkey in 2006 "involves a substitution in one sample of an amino acid at position 223 of the haemoagglutinin receptor protein. This protein allows the flu virus to bind to the receptors on the surface of its host's cells. This mutation has been observed twice before — in a father and son in Hong Kong in 2003, and in one fatal case in Vietnam last year. It increases the virus's ability to bind to human receptors, and decreases its affinity for poultry receptors, making strains with this mutation better adapted to infecting humans."[according to whom?] Another mutation in the same sample at position 153 has as yet unknown effects.[9] "Amino acid residues at positions 226 and 228 of the receptor binding pocket of HA appear to determine binding affinity to cell surface receptors and to influence the selective binding of the virus to avian (sialic acid -2,3-NeuAcGal) or human (sialic acid -2,6-NeuAcGal) cell surface receptors. The human A/HK/212/03 and A/HK/213/03 isolates retain the signature associated with avian receptor binding, but they have a unique amino acid substitution (Ser227Ile) within the receptor binding pocket that was not present even in the closely related A/Gs/HK/739.2/02 (genotype Z+) virus."[10] Recent research reveals that humans have avian type receptors at very low densities and chickens have human type receptors at very low densities.[11] Researchers "found that the mutations at two places in the gene, identified as 182 and 192, allow the virus to bind to both bird and human receptors."[12][13] See research articles Host Range Restriction and Pathogenicity in the Context of Influenza Pandemic (Centers for Disease Control and Prevention, 2006) (by Gabriele Neumann and Yoshihiro Kawaoka) and Structure and Receptor Specificity of the Hemagglutinin from an H5N1 Influenza Virus (American Association for the Advancement of Science, 2006) (by James Stevens, Ola Blixt, Terrence M. Tumpey, Jeffery K. Taubenberger, James C. Paulson, Ian A. Wilson) for further details.
- NA codes for neuraminidase which is an antigenic glycoprotein enzyme found on the surface of the influenza viruses. It helps the release of progeny viruses from infected cells. Flu drugs Tamiflu and Relenza work by inhibiting some strains of neuraminidase. They were developed based on N2 and N9. "In the N1 form of the protein, a small segment called the 150-loop is inverted, creating a hollow pocket that does not exist in the N2 and N9 proteins. [...] When the researchers looked at how existing drugs interacted with the N1 protein, they found that, in the presence of neuraminidase inhibitors, the loop changed its conformation to one similar to that in the N2 and N9 proteins."[14]
Internal encoding gene segments[]
- Internal viral protein encoding gene segments (RNA molecule): (M, NP, NS, PA, PB1, PB2) [15]
Matrix encoding gene segments[]
- M codes for the matrix proteins (M1 and M2) that, along with the two surface proteins (hemagglutinin and neuraminidase), make up the capsid (protective coat) of the virus. It encodes by using different reading frames from the same RNA segment.
- M1 is a protein that binds to the viral RNA.
- M2 is a protein that uncoats the virus, thereby exposing its contents (the eight RNA segments) to the cytoplasm of the host cell. The M2 transmembrane protein is an ion channel required for efficient infection.[16] The amino acid substitution (Ser31Asn) in M2 some H5N1 genotypes is associated with amantadine resistance.[17]
Nucleoprotein encoding gene segments.[]
- NP codes for nucleoprotein.
- NS: NS codes for two nonstructural proteins (NS1 and NS2 - formerly called ). "[T]he pathogenicity of influenza virus was related to the nonstructural (NS) gene of the H5N1/97 virus".[18]
- NS1: Non-structural: nucleus; effects on cellular RNA transport, splicing, translation. Anti-interferon protein.[19] The "NS1 of the highly pathogenic avian H5N1 viruses circulating in poultry and waterfowl in Southeast Asia might be responsible for an enhanced proinflammatory cytokine response (especially TNFa) induced by these viruses in human macrophages".[4] H5N1 NS1 is characterized by a single amino acid change at position 92. By changing the amino acid from glutamic acid to aspartic acid, the researchers were able to abrogate the effect of the H5N1 NS1. [This] single amino acid change in the NS1 gene greatly increased the pathogenicity of the H5N1 influenza virus."[20]
- NEP: The "nuclear export protein (NEP, formerly referred to as the NS2 protein) mediates the export of vRNPs".[21]
Polymerase encoding gene segments[]
- PA codes for the which is a critical component of the viral polymerase.
- PB1 codes for the and the .
- The PB1 protein is a critical component of the viral polymerase.
- The PB1-F2 protein is encoded by an alternative open reading frame of the PB1 RNA segment and "interacts with 2 components of the mitochondrial permeability transition pore complex, ANT3 and VDCA1, [sensitizing] cells to apoptosis. [...] PB1-F2 likely contributes to viral pathogenicity and might have an important role in determining the severity of pandemic influenza."[4] This was discovered by Chen et al. and reported in Nature.[5] "After comparing viruses from the Hong Kong 1997 H5N1 outbreak, one amino acid change (N66S) was found in the PB1-F2 sequence at position 66 that correlated with pathogenicity. This same amino acid change (N66S) was also found in the PB1-F2 protein of the 1918 pandemic A/Brevig Mission/18 virus."[6]
- PB2 codes for the which is a critical component of the viral polymerase. As of 2005, 75% of H5N1 human virus isolates from Vietnam had a mutation consisting of Lysine at residue 627 in the PB2 protein; which is believed to cause high levels of virulence.[22] Until H5N1, all known avian influenza viruses had a Glu at position 627, while all human influenza viruses had a lysine. As of 2007, "The emergence of 3 (or more) substrains from the EMA [EMA=Europe, Middle East, Africa] clade represents multiple new opportunities for avian influenza (H5N1) to evolve into a human pandemic strain. In contrast to strains circulating in Southeast Asia, EMA viruses are derived from a progenitor that has the PB2 627K mutation. These viruses are expected to have enhanced replication characteristics in mammals, and indeed the spread of EMA has coincided with the rapid appearance of cases in mammals—including humans in Turkey, Egypt, Iraq, and Djibouti, and cats in Germany, Austria, and Iraq. Unfortunately, the EMA-type viruses appear to be as virulent as the exclusively Asian strains: of 34 human infections outside of Asia through mid-2006, 15 have been fatal."[23] Lys at PB2–627 is believed to confer to avian H5N1 viruses the advantage of efficient growth in the upper and lower respiratory tracts of mammals.[24]
Mutation[]
Influenza viruses have a relatively high mutation rate that is characteristic of RNA viruses. The segmentation of the influenza genome facilitates genetic recombination by segment reassortment in hosts who are infected with two different influenza viruses at the same time. H5N1 viruses can reassort genes with other strains that co-infect a host organism, such as a pig, bird, or human, and mutate into a form that can pass easily among humans. This is one of many possible paths to a pandemic.
The ability of various influenza strains to show species-selectivity is largely due to variation in the hemagglutinin genes. Genetic mutations in the hemagglutinin gene that cause single amino acid substitutions can significantly alter the ability of viral hemagglutinin proteins to bind to receptors on the surface of host cells. Such mutations in avian H5N1 viruses can change virus strains from being inefficient at infecting human cells to being as efficient in causing human infections as more common human influenza virus types.[25] This doesn't mean that one amino acid substitution can cause a pandemic, but it does mean that one amino acid substitution can cause an avian flu virus that is not pathogenic in humans to become pathogenic in humans.
H3N2 ("swine flu") is endemic in pigs in China, and has been detected in pigs in Vietnam, increasing fears of the emergence of new variant strains. The dominant strain of annual flu virus in January 2006 was H3N2, which is now resistant to the standard antiviral drugs amantadine and rimantadine. The possibility of H5N1 and H3N2 exchanging genes through reassortment is a major concern. If a reassortment in H5N1 occurs, it might remain an H5N1 subtype, or it could shift subtypes, as H2N2 did when it evolved into the Hong Kong Flu strain of H3N2.
Both the H2N2 and H3N2 pandemic strains contained avian influenza virus RNA segments. "While the pandemic human influenza viruses of 1957 (H2N2) and 1968 (H3N2) clearly arose through reassortment between human and avian viruses, the influenza virus causing the 'Spanish flu' in 1918 appears to be entirely derived from an avian source".[26]
In July 2004, researchers led by H. Deng of the Harbin Veterinary Research Institute, Harbin, China and Professor Robert G. Webster of the St. Jude Children's Research Hospital, Memphis, Tennessee, reported results of experiments in which mice had been exposed to 21 isolates of confirmed H5N1 strains obtained from ducks in China between 1999 and 2002. They found "a clear temporal pattern of progressively increasing pathogenicity".[27] Results reported by Dr. Webster in July 2005 reveal further progression toward pathogenicity in mice and longer virus shedding by ducks.
Asian lineage HPAI A(H5N1) is divided into two antigenic clades. "Clade 1 includes human and bird isolates from Vietnam, Thailand, and Cambodia and bird isolates from Laos and Malaysia. Clade 2 viruses were first identified in bird isolates from China, Indonesia, Japan, and South Korea before spreading westward to the Middle East, Europe, and Africa. The clade 2 viruses have been primarily responsible for human H5N1 infections that have occurred during late 2005 and 2006, according to WHO. Genetic analysis has identified six subclades of clade 2, three of which have a distinct geographic distribution and have been implicated in human infections: Map
- Subclade 1, Indonesia
- Subclade 2, Europe, Middle East, and Africa (called EMA)
- Subclade 3, China"[28][29][30]
A 2007 study focused on the EMA subclade has shed further light on the EMA mutations. "The 36 new isolates reported here greatly expand the amount of whole-genome sequence data available from recent avian influenza (H5N1) isolates. Before our project, GenBank contained only 5 other complete genomes from Europe for the 2004–2006 period, and it contained no whole genomes from the Middle East or northern Africa. Our analysis showed several new findings. First, all European, Middle Eastern, and African samples fall into a clade that is distinct from other contemporary Asian clades, all of which share common ancestry with the original 1997 Hong Kong strain. Phylogenetic trees built on each of the 8 segments show a consistent picture of 3 lineages, as illustrated by the HA tree shown in Figure 1. Two of the clades contain exclusively Vietnamese isolates; the smaller of these, with 5 isolates, we label V1; the larger clade, with 9 isolates, is V2. The remaining 22 isolates all fall into a third, clearly distinct clade, labeled EMA, which comprises samples from Europe, the Middle East, and Africa. Trees for the other 7 segments display a similar topology, with clades V1, V2, and EMA clearly separated in each case. Analyses of all available complete influenza (H5N1) genomes and of 589 HA sequences placed the EMA clade as distinct from the major clades circulating in People's Republic of China, Indonesia, and Southeast Asia."[23]
See https://web.archive.org/web/20090709040039/http://who.int/csr/disease/avian_influenza/H5CompleteTree.pdf for a Genetic Tree of 1,342 H5N1 viruses based on their HA gene, showing their clade designations.
See also[]
References[]
- ^ Jump up to: a b c The World Health Organization Global Influenza Program Surveillance Network (2005). "Evolution of H5N1 viruses in Asia". Emerging Infectious Diseases. 11 (10): 1515–1526. doi:10.3201/eid1110.050644. PMC 3366754. PMID 16318689. Figure 1 of the article gives a diagramatic representation of the genetic relatedness of Asian H5N1 hemagglutinin genes from various isolates of the virus.
- ^ WHO (October 28, 2005). "H5N1 avian influenza: timeline" (PDF). Archived from the original (PDF) on July 27, 2011.
- ^ Canadian Press article Study shows hybrids of bird flu and human flu viruses fit well, could occur published June 1, 2008 Archived June 5, 2008, at the Wayback Machine
- ^ Jump up to: a b c García-Sastre A (January 2006). "Antiviral response in pandemic influenza viruses". Emerging Infect. Dis. 12 (1): 44–7. doi:10.3201/eid1201.051186. PMC 3291409. PMID 16494716.
- ^ Jump up to: a b c Weisan Chen; Paul A. Calvo; et al. (2001). "A novel influenza A virus mitochondrial protein that induces cell death". Nature Medicine. 7 (12): 1306–12. doi:10.1038/nm1201-1306. PMID 11726970. S2CID 723233.
- ^ Jump up to: a b Scientist Live Archived 2007-10-20 at the Wayback Machine article A Single Mutation in the PB1-F2 of H5N1 (HK/97) and 1918 Influenza A Viruses Contributes to Increased Virulence published October 18, 2007
- ^ M. T. Michael Lee, Konrad Bishop, Liz Medcalf, Debra Elton, Paul Digard, and Laurence Tiley (January 15, 2002). "Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase". Nucleic Acids Research. 30 (2): 429–438. doi:10.1093/nar/30.2.429. PMC 99831. PMID 11788704.CS1 maint: multiple names: authors list (link)
- ^ Alex Greninger (July 16, 2004). "The Definition and Measurement of Dangerous Research" (PDF). CISSM Working Paper. Archived from the original (PDF) on November 8, 2006. Retrieved 2006-12-09.
- ^ Butler D (January 2006). "Alarms ring over bird flu mutations". Nature. 439 (7074): 248–9. Bibcode:2006Natur.439..248B. doi:10.1038/439248a. PMID 16421529.
- ^ H5N1 influenza: A protean pandemic threat
- ^ Influenza Report 2006 Online book page 51
- ^ CIDRAP article Study finds 2 mutations that help H5N1 infect humans published November 21, 2006
- ^ Bloomberg News articles Two Bird Flu Gene Mutations Might Lead to Faster Human Spread published November 15, 2006
- ^ Scidev.net News article Bird flu protein's 'pocket' could inspire better drugs published August 16, 2006
- ^ The Threat of Pandemic Influenza: Are We Ready? Page 118
- ^ Influenza virus replication in Medical Microbiology, 4th edition edited by Samuel Baron. 1996 Chapter 58. ISBN 0-9631172-1-1.
- ^ Guan Y, Poon LL, Cheung CY, et al. (May 2004). "H5N1 influenza: a protean pandemic threat". Proc. Natl. Acad. Sci. U.S.A. 101 (21): 8156–61. Bibcode:2004PNAS..101.8156G. doi:10.1073/pnas.0402443101. PMC 419573. PMID 15148370.
- ^ Lee CW, Suarez DL, Tumpey TM, et al. (March 2005). "Characterization of highly pathogenic H5N1 avian influenza A viruses isolated from South Korea". J. Virol. 79 (6): 3692–702. doi:10.1128/JVI.79.6.3692-3702.2005. PMC 1075707. PMID 15731263.
Also, Pandemic Influenza Archived 2010-01-06 at the Wayback Machine Center for Infectious Disease Research & Policy Academic Health Center -- University of Minnesota - ^ NS1 described in Inhibition by the NS1 protein - Enhanced virulence/viral pathogenesis by enabling the virus to disarm the host cell type IFN defense system Pathobiologics International
- ^ "The Definition and Measurement of Dangerous Research by Alex Greninger" (PDF). Archived from the original (PDF) on 2006-03-06. Retrieved 2006-04-10.
- ^ Paragas J, Talon J, O'Neill RE, Anderson DK, García-Sastre A, Palese P (August 2001). "Influenza B and C virus NEP (NS2) proteins possess nuclear export activities". J. Virol. 75 (16): 7375–83. doi:10.1128/JVI.75.16.7375-7383.2001. PMC 114972. PMID 11462009.
- ^ The Threat of Pandemic Influenza: Are We Ready? Page 126
- ^ Jump up to: a b Salzberg SL, Kingsford C, Cattoli G, et al. (May 2007). "Genome analysis linking recent European and African influenza (H5N1) viruses". Emerging Infect. Dis. 13 (5): 713–8. doi:10.3201/eid1305.070013. PMC 2432181. PMID 17553249.
- ^ Scientist Live Archived 2007-10-18 at the Wayback Machine article Growth of H5N1 Influenza A Viruses in the Upper Respiratory Tracts of Mice published October 18, 2007
- ^ Gambaryan A, Tuzikov A, Pazynina G, Bovin N, Balish A, Klimov A (2006). "Fatal Evolution of the receptor binding phenotype of influenza A (H5) viruses". Virology. 344 (2): 432–438. doi:10.1016/j.virol.2005.08.035. PMID 16226289.
- ^ Harder, T. C.; Werner, O. (2006). "Avian Influenza". In Kamps, B. S.; Hoffman, C.; Preiser, W. (eds.). Influenza Report 2006. Paris, France: Flying Publisher. ISBN 3-924774-51-X. Retrieved 2006-04-18.
This e-book is under constant revision and is an excellent guide to Avian Influenza - ^ Chen H, Deng G, Li Z, et al. (July 2004). "The evolution of H5N1 influenza viruses in ducks in southern China". Proc. Natl. Acad. Sci. U.S.A. 101 (28): 10452–7. Bibcode:2004PNAS..10110452C. doi:10.1073/pnas.0403212101. PMC 478602. PMID 15235128.
- ^ Webster RG, Govorkova EA (November 2006). "H5N1 Influenza — Continuing Evolution and Spread". N. Engl. J. Med. 355 (21): 2174–7. doi:10.1056/NEJMp068205. PMID 17124014.
- ^ "WHO changes H5N1 strains for pandemic vaccines, raising concern over virus evolution". CIDRAP. August 18, 2006.
- ^ "Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as pre-pandemic vaccines" (PDF). WHO. August 18, 2006. Archived from the original (PDF) on August 24, 2006.
Further reading[]
- Influenza Report 2006 Online book.
- Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ (November 2005). "Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution". Nature. 437 (7062): 1162–1166. Bibcode:2005Natur.437.1162G. doi:10.1038/nature04239. PMID 16208317. presents a summary of what has been discovered in the Influenza Genome Sequencing Project.
- Links and descriptions to abstracts and full texts This bibliography of avian influenza publications was compiled through the cooperative effort of the USGS National Wildlife Health Center and the Wildlife Disease Information Node.
- Search for research publications about H5N1: Entez PubMed
- Beigel JH, Farrar J, Han AM, et al.; Writing Committee of the World Health Organization (WHO) Consultation on Human Influenza A/H5. (September 2005). "Avian influenza A (H5N1) infection in humans". N. Engl. J. Med. 353 (13): 1374–85. doi:10.1056/NEJMra052211. hdl:10722/45195. PMID 16192482.CS1 maint: multiple names: authors list (link)
- Evolutionary "Tree of Life" for H5N1:
- Here is the phylogenetic tree of the influenza virus hemagglutinin gene segment. Amino acid changes in three lineages (bird, pig, human) of the influenza virus hemagglutinin protein segment HA1.
- Here is the tree showing the evolution by reassortment of H5N1 from 1999 to 2004 that created the Z genotype in 2002.
- Here is the tree showing evolution by antigenic drift since 2002 that created dozens of highly pathogenic varieties of the Z genotype of avian flu virus H5N1, some of which are increasingly adapted to mammals.
- WHO (PDF) contains latest Evolutionary "Tree of Life" for H5N1 article Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as pre-pandemic vaccines published August 18, 2006
- Hiromoto Y, Yamazaki Y, Fukushima T, et al. (May 2000). "Evolutionary characterization of the six internal genes of H5N1 human influenza A virus". J. Gen. Virol. 81 (Pt 5): 1293–303. doi:10.1099/0022-1317-81-5-1293. PMID 10769072.
- Genome database Page links to the complete sequence of the Influenza A virus (A/Goose/Guangdong/1/96(H5N1)) genome.
External links[]
- Influenza Research Database – Database of influenza genomic sequences and related information.
- Influenza A virus subtype H5N1