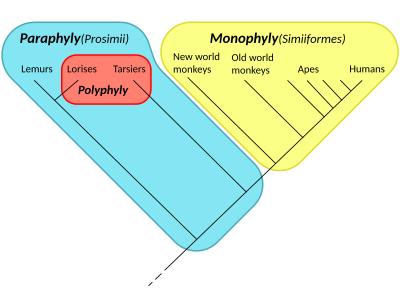
Paraphyly

In taxonomy, a group is paraphyletic if it consists of the group's last common ancestor and all descendants of that ancestor excluding a few—typically only one or two—monophyletic subgroups. The group is said to be paraphyletic with respect to the excluded subgroups. A paraphyletic group cannot be a clade, or monophyletic group, which is any group of species that includes a common ancestor and all of its descendants. One or more members of a paraphyletic group is more closely related to the excluded group(s) than it is to other members of the paraphyletic group. The term is commonly used in phylogenetics (a subfield of biology) and in linguistics. Paraphyletic groups are identified by a combination of synapomorphies and symplesiomorphies.
The term was coined by Willi Hennig to apply to well-known taxa like Reptilia (reptiles) which, as commonly named and traditionally defined, is paraphyletic with respect to mammals and birds. Reptilia contains the last common ancestor of reptiles and all descendants of that ancestor, including all extant reptiles as well as the extinct synapsids, except for mammals and birds. Other commonly recognized paraphyletic groups include fish, monkeys, and lizards.[1][page needed]
If many subgroups are missing from the named group, it is said to be polyparaphyletic.
Etymology[]
The term paraphyly, or paraphyletic, derives from the two Ancient Greek words παρά (pará), meaning "beside, near", and φῦλον (phûlon), meaning "genus, species",[2][3] and refers to the situation in which one or several monophyletic subgroups of organisms (e.g., genera, species) are left apart from all other descendants of a unique common ancestor.
Conversely, the term monophyly, or monophyletic, builds on the Ancient Greek prefix μόνος (mónos), meaning "alone, only, unique",[2][3] and refers to the fact that a monophyletic group includes organisms consisting of all the descendants of a unique common ancestor.
By comparison, the term polyphyly, or polyphyletic, uses the Ancient Greek prefix πολύς (polús), meaning "many, a lot of",[2][3] and refers to the fact that a polyphyletic group includes organisms arising from multiple ancestral sources.
Phylogenetics[]

In cladistics[]
Groups that include all the descendants of a common ancestor are said to be monophyletic. A paraphyletic group is a monophyletic group from which one or more subsidiary clades (monophyletic groups) are excluded to form a separate group. Philosopher of science Marc Ereshefsky has argued that paraphyletic taxa are the result of anagenesis in the excluded group or groups.[4] Cladists do not grant paraphyletic assemblages the status of "groups" or reify them with explanations, because they represent evolutionary non-events [5]
A group whose identifying features evolved convergently in two or more lineages is polyphyletic (Greek πολύς [polys], "many"). More broadly, any taxon that is not paraphyletic or monophyletic can be called polyphyletic. Empirically, the distinction between polyphyletic groups and paraphyletic groups is rather arbitrary, since the character states of common ancestors are inferences, not observations.
These terms were developed during the debates of the 1960s and 1970s accompanying the rise of cladistics.
Paraphyletic groupings are considered problematic by many taxonomists, as it is not possible to talk precisely about their phylogenetic relationships, their characteristic traits and literal extinction.[6][7] Related terms are stem group, chronospecies, budding cladogenesis, anagenesis, or 'grade' groupings. Paraphyletic groups are often relics from outdated hypotheses of phylogenic relationships from before the rise of cladistics.[8]
Examples[]
The prokaryotes (single-celled life forms without cell nuclei) are a paraphyletic grouping, because they exclude the eukaryotes, a descendant group. Bacteria and Archaea are prokaryotes, but archaea and eukaryotes share a common ancestor that is not ancestral to the bacteria. The prokaryote/eukaryote distinction was proposed by Edouard Chatton in 1937[9] and was generally accepted after being adopted by Roger Stanier and C.B. van Niel in 1962. The botanical code (the ICBN, now the ICN) abandoned consideration of bacterial nomenclature in 1975; currently, prokaryotic nomenclature is regulated under the ICNB with a starting date of 1 January 1980 (in contrast to a 1753 start date under the ICBN/ICN).[10]
Among plants, dicotyledons (in the traditional sense) are paraphyletic because the group excludes monocotyledons. "Dicotyledon" has not been used as a botanic classification for decades, but is allowed as a synonym of Magnoliopsida.[note 1] Phylogenetic analysis indicates that the monocots are a development from a dicot ancestor. Excluding monocots from the dicots makes the latter a paraphyletic group.[11]
Among animals, several familiar groups are not, in fact, clades. The order Artiodactyla (even-toed ungulates) as traditionally defined is paraphyletic because it excludes Cetaceans (whales, dolphins, etc.). Under the ranks of the ICZN Code, the two taxa are separate orders. Molecular studies, however, have shown that the Cetacea descend from artiodactyl ancestors, although the precise phylogeny within the order remains uncertain. Without the Cetaceans the Artiodactyls are paraphyletic.[12] The class Reptilia, as traditionally defined, is paraphyletic because it excludes birds (class Aves) and mammals. Under the ranks of the ICZN Code, these three taxa are separate classes. However, mammals hail from the synapsids (which were once described as "mammal-like reptiles") and birds are sister taxon to a group of dinosaurs (part of Diapsida), both of which are "reptiles".[13] Alternatively, reptiles are paraphyletic because they gave rise to (only) birds. Birds and reptiles together make Sauropsids, a clade of Amniota that is the sister group of the clade that includes mammals.
Osteichthyes, bony fish, are paraphyletic when circumscribed to include only Actinopterygii (ray-finned fish) and Sarcopterygii (lungfish, etc.), and to exclude tetrapods; more recently, Osteichthyes is treated as a clade, including the tetrapods.[14][15]
The "wasps" are paraphyletic, consisting of the narrow-waisted Apocrita without the ants and bees.[16] The sawflies (Symphyta) are similarly paraphyletic, forming all of the Hymenoptera except for the Apocrita, a clade deep within the sawfly tree.[14] Crustaceans are not a clade because the Hexapoda (insects) are excluded. The modern clade that spans all of them is the Tetraconata.[17][18]
One of the goals of modern taxonomy over the past fifty years has been to eliminate paraphyletic "groups" such as the examples here from formal classifications.[19][20]
Paraphyly in species[]
Species have a special status in systematics as being an observable feature of nature itself and as the basic unit of classification.[21] Some articulations of the phylogenetic species concept require species to be monophyletic, but paraphyletic species are common in nature, to the extent that they do not have a single common ancestor. Indeed, for sexually reproducing taxa, no species has a "single common ancestor" organism. Paraphyly is common in speciation, whereby a mother species (a paraspecies) gives rise to a daughter species without itself becoming extinct.[22] Research indicates as many as 20 percent of all animal species and between 20 and 50 percent of plant species are paraphyletic.[23][24] Accounting for these facts, some taxonomists argue that paraphyly is a trait of nature that should be acknowledged at higher taxonomic levels.[25][26]
Cladists advocate a phylogenetic species concept [27] that does not consider species to exhibit the properties of monophyly or paraphyly, concepts under that perspective which apply only to groups of species.[28] They consider Zander's extension of the "paraphyletic species" argument to higher taxa to represent a category error[29]
Uses for paraphyletic groups[]
When the appearance of significant traits has led a subclade on an evolutionary path very divergent from that of a more inclusive clade, it often makes sense to study the paraphyletic group that remains without considering the larger clade. For example, the Neogene evolution of the Artiodactyla (even-toed ungulates, like deer, cows, pigs and hippopotamuses - note that Cervidae, Bovidae, Suidae and Hippopotamidae, the families that contain these various artiodactyls, are all monophyletic groups) has taken place in environments so different from that of the Cetacea (whales, dolphins, and porpoises) that the Artiodactyla are often studied in isolation even though the cetaceans are a descendant group. The prokaryote group is another example; it is paraphyletic because it is composed of two Domains (Eubacteria and Archaea) and excludes (the eukaryotes). It is very useful because it has a clearly defined and significant distinction (absence of a cell nucleus, a plesiomorphy) from its excluded descendants.[citation needed]
Also, some systematists recognize paraphyletic groups as being involved in evolutionary transitions, the development of the first tetrapods from their ancestors for example. Any name given to these hypothetical ancestors to distinguish them from tetrapods—"fish", for example—necessarily picks out a paraphyletic group, because the descendant tetrapods are not included.[30] Other systematists consider reification of paraphyletic groups to obscure inferred patterns of evolutionary history.[31]
The term "evolutionary grade" is sometimes used for paraphyletic groups.[32] Moreover, the concepts of monophyly, paraphyly, and polyphyly have been used in deducing key genes for barcoding of diverse group of species.[33]
Independently evolved traits[]
Current phylogenetic hypotheses of tetrapod relationships imply that viviparity, the production of offspring without the external laying of a fertilized egg, developed independently in the lineages that led to humans (Homo sapiens) and southern water skinks (Eulampus tympanum, a kind of lizard). Put another way, viviparity is a synapomorphy for Theria within mammals, and an autapomorphy for Eulamprus tympanum (or perhaps a synapomorphy, if other Eulamprus species are also viviparous).
"Groups" based on independently-developed traits such as these examples of viviparity represent examples of polyphyly, not paraphyly.
Not paraphyly[]
- Amphibious fish are polyphyletic, not paraphyletic. Although they appear similar, several different groups of amphibious fishes such as mudskippers and lungfishes evolved independently in a process of convergent evolution in distant relatives faced with similar ecological circumstances.[34]
- Flightless birds are polyphyletic because they independently (in parallel) lost the ability to fly.[35]
- Animals with a dorsal fin are not paraphyletic, even though their last common ancestor may have had such a fin, because the Mesozoic ancestors of porpoises did not have such a fin, whereas pre-Mesozoic fish did have one.
- Quadrupedal archosaurs are not a paraphyletic group. Bipedal dinosaurs like Eoraptor, ancestral to quadrupedal ones, were descendants of the last common ancestor of quadrupedal dinosaurs and other quadrupedal archosaurs like the crocodilians.
Non-exhaustive list of paraphyletic groups[]
The following list recapitulates a number of paraphyletic groups proposed in the literature, and provides the corresponding monophyletic taxa.
Linguistics[]
The concept of paraphyly has also been applied to historical linguistics, where the methods of cladistics have found some utility in comparing languages. For instance, the Formosan languages form a paraphyletic group of the Austronesian languages because they consist of the nine branches of the Austronesian family that are not Malayo-Polynesian and are restricted to the island of Taiwan.[72]
See also[]
Notes[]
- ^ The history of flowering plant classification can be found under History of the classification of flowering plants.
References[]
- ^ Romer, A. S. (1970) [1949]. The Vertebrate Body (4th ed.). W.B. Saunders.
- ^ Jump up to: a b c Bailly, Anatole (1 January 1981). Abrégé du dictionnaire grec français. Paris: Hachette. ISBN 978-2010035289. OCLC 461974285.
- ^ Jump up to: a b c Bailly, Anatole. "Greek-french dictionary online". www.tabularium.be. Retrieved 8 March 2018.
- ^ Roberts, Keith (10 December 2007). Handbook of Plant Science. ISBN 9780470057230.
- ^ Williams, D. M. and Ebach. M. C. 2020. Cladistics: a guide to biological classification. Cambridge University Press.
- ^ Schilhab, Theresa; Stjernfelt, Frederik; Deacon, Terrence (2012). The Symbolic Species Evolved. Springer. ISBN 9789400723351.
- ^ Villmoare, Brian (2018). "Early Homo and the role of the genus in paleoanthropology". American Journal of Physical Anthropology. 165: 72–89. doi:10.1002/ajpa.23387. PMID 29380889.
- ^ Dominguez, Eduardo; Wheeler, Quentin D. (1997). "Forum – Taxonomic Stability is Ignorance". Cladistics. 13 (4): 367–372. doi:10.1111/j.1096-0031.1997.tb00325.x. S2CID 55540349.
- ^ Sapp, Jan (June 2005). "The prokaryote–eukaryote dichotomy: meanings and mythology". Microbiology and Molecular Biology Reviews. 69 (2): 292–305. doi:10.1128/MMBR.69.2.292-305.2005. PMC 1197417. PMID 15944457.
- ^ Stackebrabdt, E.; Tindell, B.; Ludwig, W.; Goodfellow, M. (1999). "Prokaryotic Diversity and Systematics". In Lengeler, Joseph W.; Drews, Gerhart; Schlegel, Hans Günter (eds.). Biology of the prokaryotes. Stuttgart: Georg Thieme Verlag. p. 679.
- ^ Jump up to: a b Simpson 2006, pp. 139–140. "It is now thought that the possession of two cotyledons is an ancestral feature for the taxa of the flowering plants and not an apomorphy for any group within. The 'dicots' ... are paraphyletic ...."
- ^ O'Leary, Maureen A. (2001). "The phylogenetic position of cetaceans: further combined data analyses, comparisons with the stratigraphic record and a discussion of character optimization". American Zoologist. 41 (3): 487–506. CiteSeerX 10.1.1.555.8631. doi:10.1093/icb/41.3.487.
- ^ Romer, A. S. & Parsons, T. S. (1985): The Vertebrate Body. (6th ed.) Saunders, Philadelphia.
- ^ Jump up to: a b c Sharkey, M. J. (2007). "Phylogeny and classification of Hymenoptera" (PDF). Zootaxa. 1668: 521–548. doi:10.11646/zootaxa.1668.1.25.
Symphyta and Apocrita have long been considered as suborders of Hymenoptera but since recognition of the paraphyletic nature of the Symphyta (Köningsmann 1977, Rasnitsyn 1988) and the advent of cladistic methods the subordinal classification should be avoided. Likewise the woodwasps are thought to be non-monophyletic, forming a grade that is ancestral relative to Apocrita and Orussidae. The traditional hymenopteran classification is faulty, by cladistic criteria,in the same way as pre-cladistic vertebrate classifications in which groups sharing plesiomorphic characterswere recognized as natural, e.g., fishes were once grouped together as 'Pisces', which excluded tetrapods.
- ^ Betancur-R, Ricardo; et al. (2013). "The Tree of Life and a New Classification of Bony Fishes". PLOS Currents Tree of Life. 5 (Edition 1). doi:10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. PMC 3644299. PMID 23653398. Archived from the original on 13 October 2013.
- ^ Johnson, Brian R.; Borowiec, Marek L.; Chiu, Joanna C.; Lee, Ernest K.; Atallah, Joel; Ward, Philip S. (2013). "Phylogenomics Resolves Evolutionary Relationships among Ants, Bees, and Wasps" (PDF). Current Biology. 23 (20): 2058–2062. doi:10.1016/j.cub.2013.08.050. PMID 24094856. S2CID 230835.
- ^ Jump up to: a b David R. Andrew (2011). "A new view of insect–crustacean relationships II. Inferences from expressed sequence tags and comparisons with neural cladistics". . 40 (3): 289–302. doi:10.1016/j.asd.2011.02.001. PMID 21315832.
- ^ Jump up to: a b Bjoern, M.; von Reumont, Ronald A.; Jenner, Matthew A.; Wills, Emiliano; Dell'Ampio, Günther; Pass, Ingo; Ebersberger, Benjamin; Meyer, Stefan; Koenemann, Thomas M. Iliffe (2012). "Pancrustacean phylogeny in the light of new phylogenomic data: support for Remipedia as the possible sister group of Hexapoda" (PDF proofs). Molecular Biology and Evolution. 29 (3): 1031–1045. doi:10.1093/molbev/msr270. PMID 22049065.
- ^ Schuh, Randall T. "The Linnaean system and its 250-year persistence." The Botanical Review 69, no. 1 (2003): 59.
- ^ Brower, Andrew V.Z. (2020). "Dead on arrival: a postmortem assessment of "phylogenetic nomenclature", 20+ years on". Cladistics. 36 (6): 627–637. doi:10.1111/cla.12432. S2CID 224927279.
- ^ Queiroz, Kevin; Donoghue, Michael J. (December 1988). "Phylogenetic Systematics and the Species Problem". Cladistics. 4 (4): 317–338. doi:10.1111/j.1096-0031.1988.tb00518.x. S2CID 40799805.
- ^ Albert, James S.; Reis, Roberto E. (8 March 2011). Historical Biogeography of Neotropical Freshwater Fishes. University of California Press. p. 308. ISBN 9780520268685. Retrieved 28 June 2011 – via Google Books.
- ^ Ross, Howard A. (July 2014). "The incidence of species-level paraphyly in animals: A re-assessment". Molecular Phylogenetics and Evolution. 76: 10–17. doi:10.1016/j.ympev.2014.02.021. PMID 24583289.
- ^ Crisp, M.D.; Chandler, G.T. (1 July 1996). "Paraphyletic species". Telopea. 6 (4): 813–844. doi:10.7751/telopea19963037. Retrieved 22 January 2015.
- ^ Zander, Richard (2013). Framework for Post-Phylogenetic Systematics. St. Louis: Zetetic Publications, Amazon CreateSpace.
- ^ Aubert, D. (2015). "A formal analysis of phylogenetic terminology: Towards a reconsideration of the current paradigm in systematics". . 66: 1–54.
- ^ Nixon, Kevin C.; Wheeler, Quentin D. (1990). "An amplification of the phylogenetic species concept". Cladistics. 6 (3): 211–23. doi:10.1111/j.1096-0031.1990.tb00541.x. S2CID 84095773.
- ^ Brower, Andrew V. Z.; Schuh, Randall T. (2021). Biological Systematics: principles and applications (3rd ed.). Ithaca, New York: Cornell University Press. ISBN 9781501752773.
- ^ Schmidt‐Lebuhn, Alexander N. (2012). "Fallacies and false premises—a critical assessment of the arguments for the recognition of paraphyletic taxa in botany". Cladistics. 28 (2): 174–87. doi:10.1111/j.1096-0031.2011.00367.x. S2CID 83900580.
- ^ Kazlev, M.A. & White, T. "Amphibians, Systematics, and Cladistics". Palaeos website. Retrieved 16 August 2012.
- ^ Patterson, Colin (1982). "Morphology and interrelationships of primitive actinopterygian fishes". American Zoologist. 22 (2): 241–259. doi:10.1093/icb/22.2.241.
- ^ Dawkins, Richard (2004). "Mammal-like Reptiles". The Ancestor's Tale, A Pilgrimage to the Dawn of Life. Boston: Houghton Mifflin. ISBN 978-0-618-00583-3.
- ^ Parhi, J.; Tripathy, P.S.; Priyadarshi, H.; Mandal S.C.; Pandey P.K. (2019). "Diagnosis of mitogenome for robust phylogeny: A case of Cypriniformes fish group". Gene. 713: 143967. doi:10.1016/j.gene.2019.143967. PMID 31279710.
- ^ Kutschera, Ulrich; Elliott, J Malcolm (26 March 2013). "Do mudskippers and lungfishes elucidate the early evolution of four-limbed vertebrates?". Evolution: Education and Outreach. 6 (8): 8. doi:10.1186/1936-6434-6-8.
- ^ Harshman, John; Braun, Edward L.; et al. (2 September 2008). "Phylogenomic evidence for multiple losses of flight in ratite birds". PNAS. 105 (36): 13462–13467. Bibcode:2008PNAS..10513462H. doi:10.1073/pnas.0803242105. PMC 2533212. PMID 18765814.
- ^ Berg, Linda (2008). Introductory Botany: Plants, People, and the Environment (2nd ed.). Belmont CA: Thomson Corporation. p. 360. ISBN 978-0-03-075453-1.
- ^ Schlegel, Martin; Hülsmann, Norbert (2 August 2007). "Protists – A textbook example for a paraphyletic taxon". Organisms Diversity & Evolution. 7 (2): 166–172. doi:10.1016/j.ode.2006.11.001. ISSN 1439-6092.
- ^ Agassiz, Louis (21 March 2013). Essay on Classification. Courier. pp. 115–. ISBN 978-0-486-15135-9.
- ^ Borchiellini, C.; Manuel, M.; Alivon, E.; Boury-Esnault, N.; Vacelet, J.; Le Parco, Y. (8 January 2001). "Sponge paraphyly and the origin of Metazoa". Journal of Evolutionary Biology. 14 (1): 171–179. doi:10.1046/j.1420-9101.2001.00244.x. PMID 29280585. S2CID 25119754.
- ^ Philippe, H; Derelle, R; Lopez, P.; et al. (April 2009). "Phylogenomics revives traditional views on deep animal relationships". Curr. Biol. 19 (8): 706–12. doi:10.1016/j.cub.2009.02.052. PMID 19345102. S2CID 15282843.CS1 maint: multiple names: authors list (link)
- ^ New data on Kimberella, the Vendian mollusc-like organism (White sea region, Russia): palaeoecological and evolutionary implications (2007), "Fedonkin, M.A.; Simonetta, A; Ivantsov, A.Y.", in Vickers-Rich, Patricia; Komarower, Patricia (eds.), The Rise and Fall of the Ediacaran Biota, Special publications, 286, London: Geological Society, pp. 157–179, doi:10.1144/SP286.12, ISBN 9781862392335, OCLC 156823511CS1 maint: uses authors parameter (link)
- ^ Butterfield, N.J. (December 2006). "Hooking some stem-group "worms": fossil lophotrochozoans in the Burgess Shale". BioEssays. 28 (12): 1161–6. doi:10.1002/bies.20507. PMID 17120226. S2CID 29130876.
- ^ Martindale, Mark; Finnerty, J.R.; Henry, J.Q. (September 2002). "The Radiata and the evolutionary origins of the bilaterian body plan". Molecular Phylogenetics and Evolution. 24 (3): 358–365. doi:10.1016/s1055-7903(02)00208-7. PMID 12220977.
- ^ Gnathifera - Richard C. Brusca
- ^ Tree of life web project – Chordates Archived 24 February 2007 at the Wayback Machine.
- ^ Tudge, Colin (2000). The Variety of Life. Oxford University Press. ISBN 0198604262.
- ^ Reeder, Tod W.; Townsend, Ted M.; Mulcahy, Daniel G.; Noonan, Brice P.; Wood, Perry L.; Sites, Jack W.; Wiens, John J. (2015). "Integrated Analyses Resolve Conflicts over Squamate Reptile Phylogeny and Reveal Unexpected Placements for Fossil Taxa". PLOS ONE. 10 (3): e0118199. Bibcode:2015PLoSO..1018199R. doi:10.1371/journal.pone.0118199. PMC 4372529. PMID 25803280.
- ^ Kielan-Jaworowska, Z. & Hurum, J. (2001). "Phylogeny and Systematics of Multituberculate Animals". Palaeontology. 44 (3): 389–429. doi:10.1111/1475-4983.00185.
- ^ Benton, Michael J. (2004). Vertebrate palaeontology (3rd ed.). Oxford: Blackwell Science. ISBN 978-0-632-05637-8.
- ^ O'Leary, Maureen A. (2001). "The Phylogenetic Position of Cetaceans: Further Combined Data Analyses, Comparisons with the Stratigraphic Record and a Discussion of Character Optimization". American Zoologist. 41 (3): 487–506. doi:10.1093/icb/41.3.487.
- ^ Savage, R. J. G. & Long, M. R. (1986). Mammal Evolution: an illustrated guide. New York: Facts on File. pp. 208. ISBN 0-8160-1194-X.
- ^ Thewissen, J. G. M.; Williams, E. M. (2002). "The Early Radiations of Cetacea (Mammalia): Evolutionary Pattern and Developmental Correlations". Annual Review of Ecology and Systematics. 33: 73–90. doi:10.1146/annurev.ecolsys.33.020602.095426. OCLC 4656321698.
- ^ Groves, C. P. (1998). "Systematics of tarsiers and lorises". Primates. 39 (1): 13–27. doi:10.1007/BF02557740. S2CID 10869981.
- ^ Johnson, Brian R.; Borowiec, Marek L.; Chiu, Joanna C.; Lee, Ernest K.; Atallah, Joel; Ward, Philip S. (2013). "Phylogenomics Resolves Evolutionary Relationships among Ants, Bees, and Wasps" (PDF). Current Biology. 23 (20): 2058–2062. doi:10.1016/j.cub.2013.08.050. PMID 24094856. S2CID 230835.
- ^ Johnson, B.R.; et al. (2013). "Phylogenomics Resolves Evolutionary Relationships among Ants, Bees, and Wasps". Current Biology. 23 (20): 2058–2062. doi:10.1016/j.cub.2013.08.050. PMID 24094856.
- ^ Parasitic Hymenoptera (Parasitica). RL Zuparko, Encyclopedia of Entomology, 2004
- ^ Lindgren, A. R.; Giribet, G.; Nishiguchi, M. K. (2004). "A combined approach to the phylogeny of Cephalopoda (Mollusca)". Cladistics. 20 (5): 454–486. doi:10.1111/j.1096-0031.2004.00032.x. S2CID 85975284.
- ^ Jump up to: a b Becker, B.; Marin, B. (2009). "Streptophyte algae and the origin of embryophytes". Annals of Botany. 103 (7): 999–1004. doi:10.1093/aob/mcp044. PMC 2707909. PMID 19273476.
- ^ Cox, Cymon J.; Li, Blaise; Foster, Peter G.; Embley, T. Martin & Civáň, Peter (2014). "Conflicting Phylogenies for Early Land Plants are Caused by Composition Biases among Synonymous Substitutions". Systematic Biology. 63 (2): 272–279. doi:10.1093/sysbio/syt109. PMC 3926305. PMID 24399481.
- ^ Christenhusz, M.J.M.; Reveal, J.L.; Farjon, A.; Gardner, M.F.; Mill, R.R.; Chase, M.W. (2011). "A new classification and linear sequence of extant gymnosperms" (PDF). Phytotaxa. 19: 55–70. doi:10.11646/phytotaxa.19.1.3.
- ^ Scoble, M.J. (1995). The Lepidoptera: form, function and diversity. Oxford: Oxford University Press. p. 404.
- ^ Stampar, S.N.; Maronna, M.M.; Kitahara, M.V.; Reimer, J.D.; Morandini, A.C. (2014). "Fast-Evolving Mitochondrial DNA in Ceriantharia: A Reflection of Hexacorallia Paraphyly?". PLOS ONE. 9 (1): e86612. Bibcode:2014PLoSO...986612S. doi:10.1371/journal.pone.0086612. PMC 3903554. PMID 24475157.
- ^ Ruppert, Edward E.; Fox, Richard, S.; Barnes, Robert D. (2004). Invertebrate Zoology, 7th edition. Cengage Learning. pp. 132–148. ISBN 978-81-315-0104-7.
- ^ Zou, H.; Zhang, J.; Li, W.; Wu, S.; Wang, G. (2012). "Mitochondrial Genome of the Freshwater Jellyfish Craspedacusta sowerbyi and Phylogenetics of Medusozoa". PLOS ONE. 7 (12): e51465. Bibcode:2012PLoSO...751465Z. doi:10.1371/journal.pone.0051465. PMC 3519871. PMID 23240028.
- ^ Marques, Antonio C.; Allen G. Collins (March 2004). "Cladistic analysis of Medusozoa and cnidarian evolution". Invertebrate Biology. 123 (1). pp. 23–42. doi:10.1111/j.1744-7410.2004.tb00139.x.
- ^ Zapata; et al. (2015). "Phylogenomic analyses support traditional relationships within Cnidaria". PLOS ONE. 10 (10): e0139068. Bibcode:2015PLoSO..1039068Z. doi:10.1371/journal.pone.0139068. PMC 4605497. PMID 26465609.
- ^ Dunn, CW; Hejnol, A; Matus, DQ; Pang, K; Browne, WE; Smith, SA; Seaver, E; Rouse, GW; et al. (2008). "Broad phylogenomic sampling improves resolution of the animal tree of life". Nature. 452 (7188): 745–749. Bibcode:2008Natur.452..745D. doi:10.1038/nature06614. PMID 18322464. S2CID 4397099.
- ^ Webster, Bonnie L.; Copley, Richard R.; Jenner, Ronald A.; Mackenzie-Dodds, Jacqueline A.; Bourlat, Sarah J.; Rota-Stabelli, Omar; Littlewood, D. T. J.; Telford, Maximilian J. (November 2006). "Mitogenomics and phylogenomics reveal priapulid worms as extant models of the ancestral Ecdysozoan". Evolution & Development. 8 (6): 502–510. doi:10.1111/j.1525-142X.2006.00123.x. PMID 17073934. S2CID 22823313.
- ^ Ruppert, Edward E.; Fox, Richard S & Barnes, Robert D. (2004), Invertebrate zoology : a functional evolutionary approach (7th ed.), Belmont, CA: Thomson-Brooks/Cole, ISBN 978-0-03-025982-1, p. 788ff. – see particularly p. 804
- ^ Shimek, Ronald (January 2006). "Nano-Animals, Part I: Rotifers". Reefkeeping.com. Retrieved 27 July 2008.
- ^ Wilson, Don E. & Reeder, DeeAnn M. (2005), Mammal Species of the World: A Taxonomic and Geographic Reference, Volume 1 (3rd ed.), Belmont, CA: Johns Hopkins University Press, ISBN 0-8018-8221-4, p. 699.
- ^ Greenhill, Simon J. and Russell D. Gray. (2009.) "Austronesian Language and Phylogenies: Myths and Misconceptions About Bayesian Computational Methods," in Austronesian Historical Linguistics and Culture History: a Festschrift for Robert Blust, edited by Alexander Adelaar and Andrew Pawley. Canberra: Pacific Linguistics, Research School of Pacific and Asian Studies, The Australian National University.
Bibliography[]
- Simpson, Michael George (2006). Plant systematics. Burlington; San Diego; London: Elsevier Academic Press. ISBN 978-0-12-644460-5.
- Paraphyletic groups as natural units of biological classification
External links[]
| Look up paraphyletic in Wiktionary, the free dictionary. |
- Funk, D. J.; Omland, K. E. (2003). "Species-level paraphyly and polyphyly: Frequency, cause and consequences, with insights from animal mitochondrial DNA" (PDF). Annual Review of Ecology, Evolution, and Systematics. 34: 397–423. doi:10.1146/annurev.ecolsys.34.011802.132421.
- Phylogenetics
- Paraphyletic groups