Proneural genes
Proneural genes encode transcription factors of the basic helix-loop-helix (bHLH) class which are responsible for the development of neuroectodermal progenitor cells. Proneural genes have multiple functions in neural development. They integrate positional information and contribute to the specification of progenitor-cell identity.[1] From the same ectodermal cell types, neural or epidermal cells can develop based on interactions between proneural and neurogenic genes.[2] Neurogenic genes are so called because loss of function mutants show an increase number of developed neural precursors. On the other hand, proneural genes mutants fail to develop neural precursor cells.[3]
The proneural genes are expressed in groups of cells (proneural clusters) from which one progenitor cell – typically the one in the middle – will be singled out, leading to the formation of many different types of neurons in the central and peripheral nervous systems.[4][5] Proneural genes encode a group of bHLH proteins that play crucial roles in controlling cell fate in a variety of tissue types.[6] Basic helix-loop-helix proteins are characterized by two alpha helixes separated by a loop. The helixes mediate dimerization, and the adjacent basic region is required for DNA binding.[1] The human genome contains approximately 125 bHLH factors.[7]
Discovery[]
The proneural genes were first identified in the 1920s[by whom?], when mutant flies that lacked subsets of external sense organs or bristles were found.[8] Later on, in the 1970s, the achaete-scute complex, a complex of genes that are involved in regulating the early steps of neural development in Drosophila, were identified[by whom?].[9] Using molecular tools it was possible to isolate the first four genes of this complex: achaete (ac), scute (sc), lethal of scute (lsc) and asense (ase).[9] Another proneural gene, atonal (ato)[10] was isolated more recently[when?] and two ato-related genes, amos and cato, were later-isolated, defining a second family of proneural genes – atonal complex.[11] Recently[when?], the first homologue of the fly proneural genes to be found in mammals was mash1.[4]
List of proneural genes[]
This list refers to bHLH proteins found in invertebrates and vertebrates. They are grouped in distinct families on the basis of closer sequence similarities in the bHLH domain:[1]
| Organisms | E-proteins | atonal family | nato family | oligo family | neuroD family | Neurogenin family | achate-scute family | Nscl family |
|---|---|---|---|---|---|---|---|---|
| invertebrates | Daughterless | Atonal, Amos, Cato, Lin32 | Nato3 | Biparous | CnASH, Achaete (ac), Asense (ase), Scute (sc), Lethal of scute (lsc) | |||
| vertebrates | E-12 | Math1, Math5 | Nato3 | Beta3, Beta4, Olig1, Olig2, Olig3 | NeuroD1 (NeuroD), NeuroD2 (Ndrf), NeuroD4 (Math3), NeuroD6 (Math2, Nex1) | amphioxNgn, Ngn1, Ngn2, Ngn3 | Cash4, Mash1, Mash2, Xash3 | Nscl1, Nscl2 |
Proneural genes functions[]
Genes of the ASC and Neurogenin families, and possibly members of the family of ato homologues, have a similar proneural function in vertebrates to that of their Drosophila counterparts, whereas other neural bHLH genes are involved in specifying neuronal fates or in neuronal differentiation, but have no proneural role.[1]
Neural functions[]
Proneural proteins bind DNA as heterodimeric complexes that are formed by bHLH proteins or E proteins. Because heterodimerization is a prerequisite for DNA binding, factors that interfere with dimerization effectively act as passive repressors of proneural gene activity. Proneural proteins specifically bind DNA sequences that contain a core hexanucleotide motif, CANNTG, known as an E-box. The basic region and helix 1 of the bHLH domain form a long alpha-helix that is connected with the loop region to helix 2. Direct contacts between bHLH residues and DNA are responsible for the common ability of neural bHLH proteins to bind to the core E-boxsequence.[1] The cells within a cluster that express a proneural gene (called a proneural cluster) can be thought of as cells of an equivalence group. Within a proneural cluster, the cells compete with each other, such that only a subset of cells is singled out to develop into neuronal precursors. This singling out process is mediated by cell-cell interactions interpreted through the action of neurogenic genes. In neuroectoderm, neurogenic genes are required to single out cells from within proneural clusters to form neuronal precursors, leaving the remaining cells of proneural clusters to develop into epidermal cells.[12] Proneural genes may function in analogous fashions in vertebrates and invertebrates, specifically they were implicated in early neurogenesis.[13] Although proneural proteins are responsible for trigger neurogenesis, different proteins are required for different neural and/or glial cell types. This implies that each of these proteins is capable of regulating both common target genes for neurogenesis and unique target genes for neuronal subtype characteristics.[14] Proneural bHLH transcription factors, not only drive neurogenesis by activating the expression of a cascade of neuronal genes, but they inhibit the expression of glial genes.[15] Neural bHLH genes have different functions depending on: the sensitivity to lateral inhibition, which determines if a cell becomes epidermal or neuronal, and whether the gene is expressed in the CNS before or after the terminal mitosis.[4]
Proneural genes promote neurogenesis and inhibit gliogenesis but some neurogenic factors can regulate both of these processes, depending on the proneural genes concentration. For example, BMPs (Bone Morphogenetic Proteins) promote neurogenesis in progenitors that express high levels of Neurogenin-1 and gliogenesis in progenitors that express low levels of Neurogenin-1.[5] Gliogenesis processes depend on low concentrations or delection of proneural genes and can be accelerated depending on which proneural genes are affected.[16]
In invertebrates[]
In Drosophila, proneural genes are first expressed in quiescent ectodermal cells that have both epidermal and neuronal potential. Proneural activity results in the selection of progenitors that are committed to a neural fate but remain multipotent, with sense organ progenitors giving rise to neurons, glia and other non-neuronal cell types. Additionally, some neuroblasts of the central nervous system also generate both neurons and glia. Progenitors of the peripheral and central nervous system only begin to divide after proneural gene expression has subsided.[1]
In vertebrates[]
Proneural genes are first expressed in neuroepithelial cells that are already specified for a neural fate and are self-renewing. Proneural activity results in the generation and delamination of progenitors that are restricted to the neuronal fate and have a limited mitotic potential. In some lineages, at least, proneural genes are involved in the commitment of neural progenitors to the neuronal fate at the expense of a glial fate.[1]
In lateral inhibition process[]

Lateral inhibition is a cell-cell interaction that occurs within a proneural cluster to determine and limit the cells that give rise to neuroblast.[16] During this interaction, nascent neuroblasts express proneural genes above a determined threshold and, at the same time, they express a membrane bound ligand, called ‘’Delta’’, which binds and activate Notch receptors expressed in neighboring cells. Once Notch is activated, the activity of proneural genes decreases in these cells, probably due to the activation of genes in the ‘’enhancer of split E(spl)’’ complex, encoding in inhibitory bHLH transcription factors.[17] When inhibited, proneural genes prevent cells from becoming neural, but also reduce their levels of ‘’Delta’’. These particular interactions restrict the proneural activity to a single cell in each proneural cluster giving rise to a salt-and-pepper pattern.[2][16]
Not all proneural genes are equally sensitive to lateral inhibition. For example, in Xenopus, Chitnis and Kintner demonstrated that ‘’XASH-3’’ and NeuroD (achaete-scute complex) respond differently to lateral inhibition, which reflect different ability to activate target genes and differential susceptibility of these target genes to repression by notch.[16] Posterior studies revealed that even when Notch/Delta signaling pathway is blocked, Wnt2b is capable of inhibiting neuronal differentiation, through the downregulation of mRNA expression of multiple proneural genes and also of Notch1. With this mechanism Wnt2b maintains progenitor cells undifferentiated by attenuating the expression of proneural and neurogenic genes, preventing cells from getting into the differentiation cascade regulated by proneural genes and Notch.[18] Although notch signaling is involved in the control of proneural gene expression, positive-feedback loops are required to increase or maintain the levels of proneural genes. The transcription factors responsible for this maintenance can act through the inhibition of the notch signaling pathway in particular cells or at a post-transcriptional level, affecting proneural genes transcription and function.[1]
In neurogenesis[]
Neurogenesis in the invertebrate nervous system[]
In invertebrates, the proneural genes, particularly the members of the achaete-scute complex (AS-C) promote neurogenesis, while the neurogenic genes prevent neurogenesis and facilitate epidermal development. The formation of neuroblasts depends on the Achaete-scute complex genes – achaete (ac), scute (sc), lethal of scute (lsc) and ventral nervous system defective (‘’vnd’’). However, only ‘’vnd’’ can control this formation process because this gene activates the expression of the others.[19][20] ac, sc, lsc factors are initially expressed within the primordium of the embryonic central nervous system (neuroectoderm) in proneural clusters, from which single neuroblasts later arise.[20] Every cell of the proneural cluster shares a common neuroblasts-forming potential. The local inhibition of the remaining cells by the enlarging neuroblasts ensures that only one neuroblast arises from the proneural cluster. All cells of the cluster retain their NB forming potential, at least while the NB is enlarging, but lose this potential by the time the cell is about to divide. The patterns of expression of the proneural genes lead to different modes of neuroblasts formation in the head and trunk. Co-expression of proneural genes in brain neuroblasts is transient and varies with the developmental stage.[21]
Neurogenesis of the stomatogastric nervous system in Drosophila[]
Proneural gene expression in the neuroectodermal cells that constitutes the proneural clusters turns them competent to delaminate as neuroblasts. Although neuroblasts are the precursors of Drosophila’s central nervous system (CNS), the proneural gene expression are also involved in control specification and morphogenesis of stomatogastric nerve cell precursors. These genes are expressed and required during all phases of the stomatogastric nervous system (SNS) development to regulate the number, pattern and structural characteristics of the SNS subpopulations. The proper balance between proneural and neurogenic gene expression in the SNS placodes is involved in the control of a complex sequence of morphogenetic movements (delamination, invagination and dissociation) by which these placodes give rise to the different SNS subpopulations.[22]
Neurogenesis in the vertebrate central nervous system[]
In central nervous system not all bHLH genes are involved in neurogenesis because NeuroD and ‘’Math3/NeuroM’’ families are also involved in the neuronal-versus-glial cell fate decision. Another pro-neural family (which includes ‘’math1’’ and ‘’math5’’) is essential to the development of a small number of neural lineages whereas ‘’math1’’ have also a role in the specification interneuron identity. Cell types that depend on ‘’math1’’ expression belong to the proprioceptive sensory pathway.[1] Bertrand et al. (2002) have confirmed the proneural activity of ‘’mash1’’, ngn1 and ngn2, and possibly math1 and ‘’math5’’ in the mouse.[1] Neurogenesis in the central nervous system depends on proneural gene inhibition by Notch signaling pathway and the absence of this key regulator results in the premature differentiation of neurons. To maintain neural progenitor cells a regulatory loop takes place between neighboring cells, that involves the lateral inhibition process (see lateral inhibition).[23] In the absence of Lateral inhibition some proneural genes such as ASCL1 or "neuroG" are capable of inducing the expression of neuron-specific genes leading to the premature formation of early born-neurons.[24] Ratié and colleagues (2013) comprised that Notch proneural gene network have an important role in cell fate renewal and transition in the mouse.[23]
Neurogenesis in the vertebrate peripheral nervous system[]
In the peripheral nervous system, Ngns are involved in the determination of all cranial and spinal sensory progenitors. Proneural genes such as mash1, ngn1 and ngn2 are mainly expressed in most progenitors of spinal cord, and are also co-express in the dorsal telencephalon. Together these groups of bHLH factors promote the generation of all cerebral cortex progenitors. Mash1 is the only gene expressed in the ventral telencephalon. However, in the ventral and dorsal ends of the neural tube a different type of proneural genes is expressed, such as ngn3 and ‘’math1’’.[1]
In gliogenesis[]
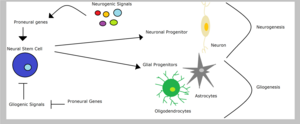
Neural stem cells could give rise to neuronal or glial progenitors, depending on the type of signals that they receive - gliogenic or neurogenic signals, respectively. Glial progenitor cells could differentiate into oligodendrocytes or astrocytes. However, lineage commitment of neural progenitors involves the suppression of alternative fates. Therefore, vertebrate proneural genes promote neuronal fates and simultaneously inhibit glial fates. For example, the downregulation of the expression of proneural gene ngn2 in the spinal cord represses oligodendrocyte differentiation. In the context of restricted glial progenitors, proneural genes might have functions that are distinct from their better-characterized role in lineage specification, perhaps in the differentiation of glial lineages.[1] Sun and colleagues showed that proneural ngn1 inhibits gliogenesis by binding transcriptional co-activators like CBP/Smad1 or p300/Smad1 preventing the transcription of glialdifferentiation genes.[15] On the other hand, the Notch signaling pathway is capable of promoting gliogenesis in stem cells through the inhibition of proneural genes, such as mash1 and neurogenins.[5]
In cell-cycle regulation fate[]
In vertebrates, although proneural genes determine the neural fate of progenitors, they also promote the arrest of their division stage by the isolation of already specified progenitor cells from the influence of extrinsic fate-determining cues. Proneural genes regulate cell cycle by the activation of cyclin-dependent kinase (‘’Cdk’’) inhibitors in some lineages at the level of neuronal-differentiation genes. On the other hand, in invertebrates like Drosophila, proneural genes are expressed mainly in non-dividing cells, but could also be expressed in dividing-cells, where Achaete-scute complex proneural genes have been shown to inhibit cell-cycle progression.[1]
In the development of sensory organs[]
The proneural genes also have an important role in the development of distinct types of sensory organs, namely chordotonal organs (proprioceptorsthat detect mechanical and sound vibrations) and external sensory organs. Members of achaete-scute complex, such as achaete and scute, as well as ‘’atonal’’ and ‘’daughterless’’ confer to ectodermal cells the ability to become sensory mother cells (SMCs).[6][10] In the development of sensory organs there are two main phases: determination and differentiation that may not be mechanistically separable. Proneural proteins are involved in both processes, through the activation of the downstream “differentiating genes”[25] that in turn regulate the induction of sensory-organ-subtype characteristics. The specification of sensory organs by proneural genes is a complex process, since they elicit different cellular contexts. For instance, in Drosophila, atonal (ato) can promote the development of chordotonal organs, for the receptors of olfactory sense organs, depending on the imaginal disc in which it is expressed.[1] In Drosophila’s embryogenesis, the proneural gene achaete is expressed in well-determined regions as in the endoderm, being responsible for the formation of particular sensory organs in the adult and larvae. According to Ruíz-Gomes and Ghysen (1993), this expression occurs in two distinct phases: a competent state, in which the proneural gene is expressed in a cell cluster; a determined state, in which a specific cell accumulate high levels of ‘’ac’’ transcripts, originating a neural precursor.[26] The function of each of the ASC complex genes varies with the development state (larvae or adult).[26] For example, in the adult state the ac and sc genes promote the differentiation of two sets of complementary sensory organs and ase gene as a minimal function, while in the larvae state ac and sc genes affect the same set of sensory organs and ase is responsible for the determination of a complementary set.[27]
In corticogenesis[]
In the neocortex exists a wide neuronal network, supported by astrocytes and oligodendrocytes (glial cells) with different functions. During cortical development, bHLH factors control proliferation and differentiation of neural cells and their functions at any given time and place depends on their cellular context. NeuroD, Ngns, Mash, ‘’Olig’’ and other proneural gene families have a crucial role in cell fate decision during corticogenesis and different combinations of them regulate the choice and the timing of differentiation into a neuron, an astrocyte or an oligodendrocyte.[28] High levels of Ngn1 and Ngn2 are required to specify neuronal identity of cortical progenitors only in early stages of neocortex development.[29] Particularly, ngn2 is also important to regulate the transition of cortical progenitors from the ventricular zone to the subventricular zone. On the other hand, mash1 is implicated in the early differentiation of striatum neurons[30] and sufficient to promote basal cell divisions independently of its role in the specification of neuronal cell fates at later stages. Cooperation between ngn2 and mash1 proneural genes regulate the transition of cortical progenitors from apical to basal cell compartments.[28] The specification of different neuronal subtypes depends on the group of proneural genes involved.[31] Low levels of proneural transcripts in ventricular zone are expressed when progenitor’s specification occurs and an increase in their expression results in the beginning of neurogenesis. Ngns are responsible for the formation of glutamatergic neurons whereas mash1 gives rise to GABAergic and cholinergic neurons.[31]
In Drosophila photoreceptor development[]
The proneural protein Atonal (Ato) is responsible for the development of Drosophila’s R8 photoreceptors. However, it does not act alone, since it dimerizes with a second identical protein, ‘’Daughterless (Da)’’ that reinforces its expression. The coexpression of ‘’Ato’’ and ‘’Da’’ is important for the migration of the different cell types of an ommatidium and for the repression of atonal in the inter-cluster spaces, functioning as inhibitory signals that regulate both the number and position of the nascent R8 cells. Moreover, this expression leads to the activation of the kinase MAPK that is important for the cellular recruitment and the repressor effect, once, when this kinase is inactive the expression of ‘’Ato’’ is detected in every cell.[32][33] In an initial state, Hedgehog (‘’Hh’’) and Decapentaplegic are responsible for the activation of ‘’ato’’ in small cellular clusters, leading to the activation of MAPK. First, Hedgehog induces ‘’ato’’ expression in more than just the cells that ultimately become photoreceptor cells, so its expression has to be refined and restricted to a single presumptive R8 cell. Hedgehog signaling is also required to repress ‘’atonal’’ expression between the nascent proneural clusters, which reveals a dual role crucial to building precision and geometry into the adult retina. Therefore, the regulation of ‘’ato’’ expression depends on the levels of Hedgehog: at low levels (that is far from the source) this pronuclear gene is activated and at high levels (close to the source) it is repressed. Combining lateral inhibition with the dual role played by Hedgehog, one can imagine how the hexagonal array of R8 cells can be patterned.[33]
Non-neural functions[]
Besides their role in the nervous system development, proneural genes are also involved in processes related to trophoblast invasion, endocrine cell differentiation (namely in pancreas and adenohypophysis), sex determination and lungs, thyroid, adrenal and salivary glands and gastrointestinal system development.[34]
In human trophoblast invasion[]
Additionally to their involvement in neuronal and glial differentiation, sex determination and sensory organs development, proneural genes are also involved in trophoblast differentiation during progression of invasion, in placental formation. Studies revealed the expression of neuroD1, neuroD2 and ‘’ath2’’ transcripts in different subsets of invasive trophoblast.[35]
In pancreas development[]
In the developing pancreas, transcription factor ngn3’marks the populations of cells that are in transit from undifferentiated epithelial progenitor cells to mature endocrine cells (precursors of pancreatic endocrine cells), and thus that do not express hormones yet, which suggests that this gene is turned off in differentiated hormone-positive.[36] ngn3’’ is both necessary and sufficient to drive the formation of islet cells during pancreatic development, in a manner similar to the specification of neural fate in neuroectoderm.[37]
In sex determination[]
Although neurogenesis and sex determination appear to be different biological processes, there is evidence that ‘’daughterless (da’’) - an essential gene for the formation of the entire Drosophila peripheral nervous system - is also required for proper sex determination.[38] In flies, scute works to direct neuronal development, but this gene also acquired a role in the primary event of sex determination – X chromosome counting – by becoming an X chromosome signal element.[39]
In myogenesis[]
Although proneural genes operate in the ectoderm, lethal of scute acts in the somatic mesoderm to define cell cluster from which muscle progenitors will be single out.[40] The interaction between these cells and ectoderm, leads to the formation of muscle founder cells, in an analogous process to the one that occurs in the central nervous system.[41]
In cell migration[]
ngn1 and ngn2 can regulate independently the mechanisms of the cell migration and are involved in the initial downregulation of RhoA right before neural progenitor cells become postmitotic, whereas neuroD is primarily involved in the continuous suppression of RhoA during the cortical migration. Other bHLH factors like ‘’math2’’, neuroD2 and ‘’nscl1’’ can suppress RhoA expression and regulate the migration machinery in postmitotic neurons.[42]
References[]
- ^ a b c d e f g h i j k l m n Bertrand, N; Castro, D. S.; Guillemot, F (2002). "Proneural genes and the specification of neural cell types". Nature Reviews Neuroscience. 3 (7): 517–30. doi:10.1038/nrn874. PMID 12094208. S2CID 205512016.
- ^ a b Kunisch, M; Haenlin, M; Campos-Ortega, J. A. (1994). "Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins". Proceedings of the National Academy of Sciences of the United States of America. 91 (21): 10139–43. Bibcode:1994PNAS...9110139K. doi:10.1073/pnas.91.21.10139. PMC 44973. PMID 7937851.
- ^ Tepass, U; Hartenstein, V (1995). "Neurogenic and proneural genes control cell fate specification in the Drosophila endoderm". Development. 121 (2): 393–405. doi:10.1242/dev.121.2.393. PMID 7768181.
- ^ a b c Brunet, J. F.; Ghysen, A (1999). "Deconstructing cell determination: Proneural genes and neuronal identity". BioEssays. 21 (4): 313–8. doi:10.1002/(SICI)1521-1878(199904)21:4<313::AID-BIES7>3.0.CO;2-C. PMID 10377893.
- ^ a b c Morrison, S. J. (2001). "Neuronal differentiation: Proneural genes inhibit gliogenesis". Current Biology. 11 (9): R349–51. doi:10.1016/S0960-9822(01)00191-9. PMID 11369245.
- ^ a b Chien, C. T.; Hsiao, C. D.; Jan, L. Y.; Jan, Y. N. (1996). "Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes". Proceedings of the National Academy of Sciences of the United States of America. 93 (23): 13239–44. Bibcode:1996PNAS...9313239C. doi:10.1073/pnas.93.23.13239. PMC 24077. PMID 8917575.
- ^ Ledent, V; Paquet, O; Vervoort, M (2002). "Phylogenetic analysis of the human basic helix-loop-helix proteins". Genome Biology. 3 (6): RESEARCH0030. doi:10.1186/gb-2002-3-6-research0030. PMC 116727. PMID 12093377.
- ^ Ghysen, A; Dambly-Chaudière, C (1988). "From DNA to form: The achaete-scute complex". Genes & Development. 2 (5): 495–501. doi:10.1101/gad.2.5.495. PMID 3290049.
- ^ a b González, F; Romani, S; Cubas, P; Modolell, J; Campuzano, S (1989). "Molecular analysis of the asense gene, a member of the achaete-scute complex of Drosophila melanogaster, and its novel role in optic lobe development". The EMBO Journal. 8 (12): 3553–62. doi:10.1002/j.1460-2075.1989.tb08527.x. PMC 402034. PMID 2510998.
- ^ a b Jarman, A. P.; Grau, Y; Jan, L. Y.; Jan, Y. N. (1993). "Atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system". Cell. 73 (7): 1307–21. doi:10.1016/0092-8674(93)90358-w. PMID 8324823.
- ^ Goulding, S. E.; White, N. M.; Jarman, A. P. (2000). "Cato encodes a basic helix-loop-helix transcription factor implicated in the correct differentiation of Drosophila sense organs". Developmental Biology. 221 (1): 120–31. doi:10.1006/dbio.2000.9677. PMID 10772796.
- ^ Jan, Y. N.; Jan, L. Y. (1993). "HLH proteins, fly neurogenesis, and vertebrate myogenesis". Cell. 75 (5): 827–30. doi:10.1016/0092-8674(93)90525-u. PMID 8252617.
- ^ Campos-Ortega, J. A. (1993). "Mechanisms of early neurogenesis in Drosophila melanogaster". Journal of Neurobiology. 24 (10): 1305–27. doi:10.1002/neu.480241005. PMID 8228961.
- ^ Powell, L. M.; Jarman, A. P. (2008). "Context dependence of proneural bHLH proteins". Current Opinion in Genetics & Development. 18 (5): 411–7. doi:10.1016/j.gde.2008.07.012. PMC 3287282. PMID 18722526.
- ^ a b Sun, Y; Nadal-Vicens, M; Misono, S; Lin, M. Z.; Zubiaga, A; Hua, X; Fan, G; Greenberg, M. E. (2001). "Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms". Cell. 104 (3): 365–76. doi:10.1016/S0092-8674(01)00224-0. PMID 11239394.
- ^ a b c d Chitnis, A; Kintner, C (1996). "Sensitivity of proneural genes to lateral inhibition affects the pattern of primary neurons in Xenopus embryos". Development. 122 (7): 2295–301. doi:10.1242/dev.122.7.2295. PMID 8681809.
- ^ Schlatter, R.; Maier, D. (2005). "The Enhancer of split and Achaete-Scute complexes of Drosophilids derived from simple ur-complexes preserved in mosquito and honeybee". BMC Evolutionary Biology. 5: 67. doi:10.1186/1471-2148-5-67. PMC 1310631. PMID 16293187.
- ^ Kubo, F; Takeichi, M; Nakagawa, S (2005). "Wnt2b inhibits differentiation of retinal progenitor cells in the absence of Notch activity by downregulating the expression of proneural genes". Development. 132 (12): 2759–70. doi:10.1242/dev.01856. PMID 15901663.
- ^ Skeath, J. B.; Panganiban, G. F.; Carroll, S. B. (1994). "The ventral nervous system defective gene controls proneural gene expression at two distinct steps during neuroblast formation in Drosophila". Development. 120 (6): 1517–24. doi:10.1242/dev.120.6.1517. PMID 8050360.
- ^ a b Skeath, J. B.; Carroll, S. B. (1992). "Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo". Development. 114 (4): 939–46. doi:10.1242/dev.114.4.939. PMID 1618155.
- ^ Urbach, R; Schnabel, R; Technau, G. M. (2003). "The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila". Development. 130 (16): 3589–606. doi:10.1242/dev.00528. PMID 12835378.
- ^ Hartenstein, V; Tepass, U; Gruszynski-Defeo, E (1996). "Proneural and neurogenic genes control specification and Morphogenesis of stomatogastric nerve cell precursors in Drosophila". Developmental Biology. 173 (1): 213–27. doi:10.1006/dbio.1996.0018. PMID 8575623.
- ^ a b Ratié, L; Ware, M; Barloy-Hubler, F; Romé, H; Gicquel, I; Dubourg, C; David, V; Dupé, V (2013). "Novel genes upregulated when NOTCH signalling is disrupted during hypothalamic development". Neural Development. 8: 25. doi:10.1186/1749-8104-8-25. PMC 3880542. PMID 24360028.
- ^ Kageyama, R.; Ohtsuka, T.; Shimojo, H.; Imayoshi, I. (2008). "Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition". Nature Neuroscience. 11 (11): 1247–1251. doi:10.1038/nn.2208. PMID 18956012. S2CID 24613095.
- ^ Cubas, P; De Celis, J. F.; Campuzano, S; Modolell, J (1991). "Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc". Genes & Development. 5 (6): 996–1008. doi:10.1101/gad.5.6.996. PMID 2044965.
- ^ a b Ruiz-Gómez, M; Ghysen, A (1993). "The expression and role of a proneural gene, achaete, in the development of the larval nervous system of Drosophila". The EMBO Journal. 12 (3): 1121–30. doi:10.1002/j.1460-2075.1993.tb05753.x. PMC 413313. PMID 8458326.
- ^ Dambly-Chaudiere, C.; Ghysen, A. (1987). "Independent subpatterns of sense organs require independent genes of the achaete-scute complex in Drosophila larvae". Genes & Development. 1 (3): 297–306. doi:10.1101/gad.1.3.297.
- ^ a b Ross, S. E.; Greenberg, M. E.; Stiles, C. D. (2003). "Basic Helix-Loop-Helix Factors in Cortical Development". Neuron. 39 (1): 13–25. doi:10.1016/S0896-6273(03)00365-9. PMID 12848929.
- ^ Schuurmans, C; Armant, O; Nieto, M; Stenman, J. M.; Britz, O; Klenin, N; Brown, C; Langevin, L. M.; Seibt, J; Tang, H; Cunningham, J. M.; Dyck, R; Walsh, C; Campbell, K; Polleux, F; Guillemot, F (2004). "Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways". The EMBO Journal. 23 (14): 2892–902. doi:10.1038/sj.emboj.7600278. PMC 514942. PMID 15229646.
- ^ Yun, K; Fischman, S; Johnson, J; Hrabe De Angelis, M; Weinmaster, G; Rubenstein, J. L. (2002). "Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon". Development. 129 (21): 5029–40. doi:10.1242/dev.129.21.5029. PMID 12397111.
- ^ a b Britz, O; Mattar, P; Nguyen, L; Langevin, L. M.; Zimmer, C; Alam, S; Guillemot, F; Schuurmans, C (2006). "A role for proneural genes in the maturation of cortical progenitor cells". Cerebral Cortex. 16 Suppl 1: i138–51. doi:10.1093/cercor/bhj168. PMID 16766700.
- ^ Chen, C. K.; Chien, C. T. (1999). "Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors". Proceedings of the National Academy of Sciences of the United States of America. 96 (9): 5055–60. Bibcode:1999PNAS...96.5055C. doi:10.1073/pnas.96.9.5055. PMC 21815. PMID 10220417.
- ^ a b Domínguez, M (1999). "Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development". Development. 126 (11): 2345–53. doi:10.1242/dev.126.11.2345. PMID 10225994.
- ^ Pogoda, H. M.; von Der Hardt, S; Herzog, W; Kramer, C; Schwarz, H; Hammerschmidt, M (2006). "The proneural gene ascl1a is required for endocrine differentiation and cell survival in the zebrafish adenohypophysis". Development. 133 (6): 1079–89. doi:10.1242/dev.02296. PMID 16481349.
- ^ Westerman, B. A.; Poutsma, A; Maruyama, K; Schrijnemakers, H. F.; Van Wijk, I. J.; Oudejans, C. B. (2002). "The proneural genes NEUROD1 and NEUROD2 are expressed during human trophoblast invasion". Mechanisms of Development. 113 (1): 85–90. doi:10.1016/S0925-4773(01)00665-7. PMID 11900979.
- ^ Johansson, K. A.; Dursun, U.; Jordan, N.; Gu, G.; Beermann, F.; Gradwohl, G. R.; Grapin-Botton, A. (2007). "Temporal Control of Neurogenin3 Activity in Pancreas Progenitors Reveals Competence Windows for the Generation of Different Endocrine Cell Types". Developmental Cell. 12 (3): 457–465. doi:10.1016/j.devcel.2007.02.010. PMID 17336910.
- ^ Lee, J. C.; Smith, S. B.; Watada, H; Lin, J; Scheel, D; Wang, J; Mirmira, R. G.; German, M. S. (2001). "Regulation of the pancreatic pro-endocrine gene neurogenin3". Diabetes. 50 (5): 928–36. doi:10.2337/diabetes.50.5.928. PMID 11334435.
- ^ Cline, T. W. (1976). "A sex-specific, temperature-sensitive maternal effect of the daughterless mutation of Drosophila melanogaster". Genetics. 84 (4): 723–42. doi:10.1093/genetics/84.4.723. PMC 1213604. PMID 827461.
- ^ Wrischnik, L. A.; Timmer, J. R.; Megna, L. A.; Cline, T. W. (2003). "Recruitment of the proneural gene scute to the Drosophila sex-determination pathway". Genetics. 165 (4): 2007–27. doi:10.1093/genetics/165.4.2007. PMC 1462923. PMID 14704182.
- ^ Carmena, A; Bate, M; Jiménez, F (1995). "Lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis". Genes & Development. 9 (19): 2373–83. doi:10.1101/gad.9.19.2373. PMID 7557389.
- ^ Bate, C. M.; Grunewald, E. B. (1981). "Embryogenesis of an insect nervous system II: A second class of neuron precursor cells and the origin of the intersegmental connectives". Journal of Embryology and Experimental Morphology. 61: 317–30. PMID 7264548.
- ^ Ge, W; He, F; Kim, K. J.; Blanchi, B; Coskun, V; Nguyen, L; Wu, X; Zhao, J; Heng, J. I.; Martinowich, K; Tao, J; Wu, H; Castro, D; Sobeih, M. M.; Corfas, G; Gleeson, J. G.; Greenberg, M. E.; Guillemot, F; Sun, Y. E. (2006). "Coupling of cell migration with neurogenesis by proneural bHLH factors". Proceedings of the National Academy of Sciences. 103 (5): 1319–24. Bibcode:2006PNAS..103.1319G. doi:10.1073/pnas.0510419103. PMC 1345712. PMID 16432194.
- Developmental genes and proteins