Molecular paleontology
| Part of a series on |
| Paleontology |
|---|
 |
|
Paleontology Portal Category |
Molecular paleontology refers to the recovery and analysis of DNA, proteins, carbohydrates, or lipids, and their diagenetic products from ancient human, animal, and plant remains.[1][2] The field of molecular paleontology has yielded important insights into evolutionary events, species' diasporas, the discovery and characterization of extinct species. By applying molecular analytical techniques to DNA in fossils, one can quantify the level of relatedness between any two organisms for which DNA has been recovered.[3]
Advancements in the field of molecular paleontology have allowed scientists to pursue evolutionary questions on a genetic level rather than relying on phenotypic variation alone. Using various biotechnological techniques such as DNA isolation, amplification, and sequencing[4] scientists have been able to gain expanded new insights into the divergence and evolutionary history of countless organisms.
In February 2021, scientists reported, for the first time, the sequencing of DNA from animal remains, a mammoth in this instance, over a million years old, the oldest DNA sequenced to date.[5][6]
History[]
The study of molecular paleontology is said to have begun with the discovery by Abelson of 360 million year old amino acids preserved in fossil shells.[7] However, Svante Pääbo is often the one considered to be the founder of the field of molecular paleontology.[8]
The field of molecular paleontology has had several major advances since the 1950s and is a continuously growing field. Below is a timeline showing notable contributions that have been made.
Timeline[]
mid-1950s: Abelson found preserved amino acids in fossil shells that were about 360 million years old. Produced idea of comparing fossil amino acid sequences with existing organism so that molecular evolution could be studied.[7]
1970s: Fossil peptides are studied by amino acid analysis.[9] Start to use whole peptides and immunological methods.[10]
Late 1970s: Palaeobotanists (can also be spelled as Paleobotanists) studied molecules from well-preserved fossil plants.[11]
1984: The first successful DNA sequencing of an extinct species, the quagga, a zebra-like species.[1]
1991: Published article on the successful extraction of proteins from the fossil bone of a dinosaur, specifically the Seismosaurus.[12]
2005: Scientists resurrect extinct 1918 influenza virus.[13]
2006: Neanderthals nuclear DNA sequence segments begin to be analyzed and published.[18]
2007: Scientists synthesize entire extinct human endogenous retrovirus (HERV-K) from scratch.[14]
2010: A new species of early hominid, the Denisovans, discovered from mitochondrial and nuclear genomes recovered from bone found in a cave in Siberia. Analysis showed that the Denisovan specimen lived approximately 41,000 years ago, and shared a common ancestor with both modern humans and Neanderthals approximately 1 million years ago in Africa.[15]
2013: The first entire Neanderthal genome is successfully sequenced. More information can be found at the Neanderthal genome project.[16]
2013: A 400,000-year-old specimen with remnant mitochondrial DNA sequenced and is found to be a common ancestor to Neanderthals and Denisovans, Homo heidelbergensis.[17]
2015: A 110,000-year-old fossil tooth containing DNA from Denisovans was reported.[19][20]
The quagga[]
The first successful DNA sequencing of an extinct species was in 1984, from a 150-year-old museum specimen of the quagga, a zebra-like species.[1] Mitochondrial DNA (also known as mtDNA) was sequenced from desiccated muscle of the quagga, and was found to differ by 12 base substitutions from the mitochondrial DNA of a mountain zebra. It was concluded that these two species had a common ancestor 3-4 million years ago, which is consistent with known fossil evidence of the species.[21]
Denisovans[]
The Denisovans of Eurasia, a hominid species related to Neanderthals and humans, was discovered as a direct result of DNA sequencing of a 41,000-year-old specimen recovered in 2008. Analysis of the mitochondrial DNA from a retrieved finger bone showed the specimen to be genetically distinct from both humans and Neanderthals. Two teeth and a toe bone were later found to belong to different individuals with the same population. Analysis suggests that both the Neanderthals and Denisovans were already present throughout Eurasia when modern humans arrived.[16] In November 2015, scientists reported finding a fossil tooth containing DNA from Denisovans, and estimated its age at 110,000-years-old.[19][20]
Mitochondrial DNA analysis[]

The mtDNA from the Denisovan finger bone differs from that of modern humans by 385 bases (nucleotides) in the mtDNA strand out of approximately 16,500, whereas the difference between modern humans and Neanderthals is around 202 bases. In contrast, the difference between chimpanzees and modern humans is approximately 1,462 mtDNA base pairs.[22] This suggested a divergence time around one million years ago. The mtDNA from a tooth bore a high similarity to that of the finger bone, indicating they belonged to the same population.[23] From a second tooth, an mtDNA sequence was recovered that showed an unexpectedly large number of genetic differences compared to that found in the other tooth and the finger, suggesting a high degree of mtDNA diversity. These two individuals from the same cave showed more diversity than seen among sampled Neanderthals from all of Eurasia, and were as different as modern-day humans from different continents.[24]
Nuclear genome analysis[]
Isolation and sequencing of nuclear DNA has also been accomplished from the Denisova finger bone. This specimen showed an unusual degree of DNA preservation and low level of contamination. They were able to achieve near-complete genomic sequencing, allowing a detailed comparison with Neanderthal and modern humans. From this analysis, they concluded, in spite of the apparent divergence of their mitochondrial sequence, the Denisova population along with Neanderthal shared a common branch from the lineage leading to modern African humans. The estimated average time of divergence between Denisovan and Neanderthal sequences is 640,000 years ago, and the time between both of these and the sequences of modern Africans is 804,000 years ago. They suggest the divergence of the Denisova mtDNA results either from the persistence of a lineage purged from the other branches of humanity through genetic drift or else an introgression from an older hominin lineage.[23]
Homo heidelbergensis[]

Homo heidelbergensis was first discovered in 1907 near Heidelberg, Germany and later also found elsewhere in Europe, Africa, and Asia.[25][26] However it was not until 2013 that a specimen with retrievable DNA was found, in a ~400,000 year old femur found in the Sima de los Huesos Cave in Spain. The femur was found to contain both mtDNA and nuclear DNA. Improvements in DNA extraction and library preparation techniques allowed for mtDNA to be successfully isolated and sequenced, however the nuclear DNA was found to be too degraded in the observed specimen, and was also contaminated with DNA from an ancient cave bear (Ursus deningeri) present in the cave.[27] The mtDNA analysis found a surprising link between the specimen and the Denisovans, and this finding raised many questions. Several scenarios were proposed in a January 2014 paper titled "A mitochondrial genome sequence of a hominin from Sima de los Huesos", elucidating the lack of convergence in the scientific community on how Homo heidelbergensis is related to other known hominin groups. One plausible scenario that the authors proposed was that the H. heidelbergensis was an ancestor to both Denisovans and Neanderthals.[27] Completely sequenced nuclear genomes from both Denisovans and Neanderthals suggest a common ancestor approximately 700,000 years ago, and one leading researcher in the field, Svante Paabo, suggests that perhaps this new hominin group is that early ancestor.[17]
Applications[]
Discovery and characterization of new species[]
Molecular paleontology techniques applied to fossils have contributed to the discovery and characterization of several new species, including the Denisovans and Homo heidelbergensis. We have been able to better understand the path that humans took as they populated the earth, and what species were present during this diaspora.
De-extinction[]

It is now possible to revive extinct species using molecular paleontology techniques. This was first accomplished via cloning in 2003 with the Pyrenean ibex, a type of wild goat that became extinct in 2000. Nuclei from the Pyrenean ibex's cells were injected into goat eggs emptied of their own DNA, and implanted into surrogate goat mothers.[28] The offspring lived only seven minutes after birth, due to defects in its lungs. Other cloned animals have been observed to have similar lung defects.[29]
There are many species that have gone extinct as a direct result of human activity. Some examples include the dodo, the great auk, the Tasmanian tiger, the Chinese river dolphin, and the passenger pigeon. An extinct species can be revived by using allelic replacement[30] of a closely related species that is still living. By only having to replace a few genes within an organism, instead of having to build the extinct species' genome from scratch, it could be possible to bring back several species in this way, even Neanderthals.[citation needed]
The ethics surrounding the re-introduction of extinct species are very controversial. Critics of bringing extinct species back to life contend that it would divert limited money and resources from protecting the world's current biodiversity problems.[31] With current extinction rates approximated to be 100 to 1,000 times the background extinction rate,[32] it is feared that a de-extinction program might lessen public concerns over the current mass extinction crisis, if it is believed that these species can simply be brought back to life. As the editors of a Scientific American article on de-extinction pose: Should we bring back the woolly mammoth only to let elephants become extinct in the meantime?[31] The main driving factor for the extinction of most species in this era (post 10,000 BC) is the loss of habitat, and temporarily bringing back an extinct species will not recreate the environment they once inhabited.[33]
Proponents of de-extinction, such as George Church, speak of many potential benefits. Reintroducing an extinct keystone species, such as the woolly mammoth, could help re-balance the ecosystems that once depended on them. Some extinct species could create broad benefits for the environments they once inhabited, if returned. For example, woolly mammoths may be able to slow the melting of the Russian and Arctic tundra in several ways such as eating dead grass so that new grass can grow and take root, and periodically breaking up the snow, subjecting the ground below to the arctic air. These techniques could also be used to reintroduce genetic diversity in a threatened species, or even introduce new genes and traits to allow the animals to compete better in a changing environment.[34]
Research and technology[]
When a new potential specimen is found, scientists normally first analyze for cell and tissue preservation using histological techniques, and test the conditions for the survivability of DNA. They will then attempt to isolate a DNA sample using the technique described below, and conduct a PCR amplification of the DNA to increase the amount of DNA available for testing. This amplified DNA is then sequenced. Care is taken to verify that the sequence matches the phylogenetic traits of the organism.[1] When an organism dies, a technique called amino acid dating can be used to age the organism. It inspects the degree of racemization of aspartic acid, leucine, and alanine within the tissue. As time passes, the D/L ratio (where "D" and "L" are mirror images of each other) increase from 0 to 1.[35] In samples where the D/L ratio of aspartic acid is greater than 0.08, ancient DNA sequences can not be retrieved (as of 1996).[36]
Mitochondrial DNA vs. nuclear DNA[]
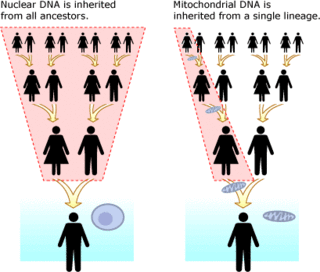
Mitochondrial DNA (mtDNA) is separate from one's nuclear DNA. It is present in organelles called mitochondria in each cell. Unlike nuclear DNA, which is inherited from both parents and rearranged every generation, an exact copy of mitochondrial DNA gets passed down from mother to her sons and daughters. The benefits of performing DNA analysis with Mitochondrial DNA is that it has a far smaller mutation rate than nuclear DNA, making tracking lineages on the scale of tens of thousands of years much easier. Knowing the base mutation rate for mtDNA,[37] (in humans this rate is also known as the Human mitochondrial molecular clock) one can determine the amount of time any two lineages have been separated. Another advantage of mtDNA is that thousands of copies of it exist in every cell, whereas only two copies of nuclear DNA exist in each cell.[38] All eukaryotes, a group which includes all plants, animals, and fungi, have mtDNA.[39] A disadvantage of mtDNA is that only the maternal line is represented. For example, a child will inherit 1/8 of its DNA from each of its eight great-grandparents, however it will inherit an exact clone of its maternal great-grandmother's mtDNA. This is analogous to a child inheriting only his paternal great-grandfather's last name, and not a mix of all of the eight surnames.
Isolation[]
There are many things to consider when isolating a substance. First, depending upon what it is and where it is located, there are protocols that must be carried out in order to avoid contamination and further degradation of the sample.[4] Then, handling of the materials is usually done in a physically isolated work area and under specific conditions (i.e. specific Temperature, moisture, etc...) also to avoid contamination and further loss of sample.[4]
Once the material has been obtained, depending on what it is, there are different ways to isolate and purify it. DNA extraction from fossils is one of the more popular practices and there are different steps that can be taken to get the desired sample.[4] DNA extracted from amber-entombed fossils can be taken from small samples and mixed with different substances, centrifuged, incubated, and centrifuged again.[40] On the other hand, DNA extraction from insects can be done by grinding the sample, mixing it with buffer, and undergoing purification through glass fiber columns.[41] In the end, regardless of how the sample was isolated for these fossils, the DNA isolated must be able to undergo amplification.[4][40][41]
Amplification[]

The field of molecular paleontology benefited greatly from the invention of the polymerase chain reaction(PCR), which allows one to make billions of copies of a DNA fragment from just a single preserved copy of the DNA. One of the biggest challenges up until this point was the extreme scarcity of recovered DNA because of degradation of the DNA over time.[1]
Sequencing[]
DNA sequencing is done to determine the order of nucleotides and genes.[42] There are many different materials from which DNA can be extracted. In animals, the mitochondrial chromosome can be used for molecular study. Chloroplasts can be studied in plants as a primary source of sequence data.[42]

In the end, the sequences generated are used to build evolutionary trees.[42] Methods to match data sets include: maximum probability, minimum evolution (also known as neighbor-joining) which searches for the tree with shortest overall length, and the maximum parsimony method which finds the tree requiring the fewest character-state changes.[42] The groups of species defined within a tree can also be later evaluated by statistical tests, such as the bootstrap method, to see if they are indeed significant.[42]
Limitations and challenges[]
Ideal environmental conditions for preserving DNA where the organism was desiccated and uncovered are difficult to come by, as well as maintaining their condition until analysis. Nuclear DNA normally degrades rapidly after death by endogenous hydrolytic processes,[36] by UV radiation,[1] and other environmental stressors.
Also, interactions with the organic breakdown products of surrounding soil have been found to help preserve biomolecular materials.[43] However, they have also created the additional challenge of being able to separate the various components in order to be able to conduct the proper analysis on them.[44] Some of these breakdowns have also been found to interfere with the action of some of the enzymes used during PCR.[43]
Finally, one of the largest challenge in extracting ancient DNA, particularly in ancient human DNA, is in contamination during PCR. Small amounts of human DNA can contaminate the reagents used for extraction and PCR of ancient DNA. These problems can be overcome by rigorous care in the handling of all solutions as well as the glassware and other tools used in the process. It can also help if only one person performs the extractions, to minimize different types of DNA present.[36]
See also[]
- Ancient DNA
- Ancient protein
- Archaeogenetics
- Fossils
- Human mitochondrial DNA haplogroups
- Human Y-chromosome DNA haplogroup
- Models of DNA Evolution
- Molecular evolution
- Paleobiochemistry
- Paleobiology
- Paleobotany
References[]
- ^ Jump up to: a b c d e f g Marota, Isolina; Franco Rollo (2002). "Molecular paleontology". Cellular and Molecular Life Sciences. 59 (1): 97–111. doi:10.1007/s00018-002-8408-8. PMID 11846037. S2CID 6377604.
- ^ Higby Schweitzer, Mary (April 2004). "Molecular paleontology: some current advances and problems". Annales de Paléontologie. 90 (2): 81–102. doi:10.1016/j.annpal.2004.02.001. Retrieved 22 April 2014.
- ^ Shapiro, B.; Hofreiter, M. (23 January 2014). "A Paleogenomic Perspective on Evolution and Gene Function: New Insights from Ancient DNA". Science. 343 (6169): 1236573. doi:10.1126/science.1236573. PMID 24458647. S2CID 1015148.
- ^ Jump up to: a b c d e Waggoner, Ben. "Molecular Palaeontology" (PDF). Nature Publishing Group.
- ^ Hunt, Katie (17 February 2021). "World's oldest DNA sequenced from a mammoth that lived more than a million years ago". CNN News. Retrieved 17 February 2021.
- ^ Callaway, Ewen (17 February 2021). "Million-year-old mammoth genomes shatter record for oldest ancient DNA - Permafrost-preserved teeth, up to 1.6 million years old, identify a new kind of mammoth in Siberia". Nature. 590 (7847): 537–538. doi:10.1038/d41586-021-00436-x. PMID 33597786.
- ^ Jump up to: a b c Abelson, PH (1954). "Organic constituents of fossils". Carnegie Institute of Washington Yearbook. 53: 97–101.
- ^ Hreha, Sarah. "2013 Gruber Genetics Prize Press Release". Gruber Foundation.
- ^ Jump up to: a b de Jong, EW; Westbroek P; Westbroek JF; Bruining JW (1974). "Preservation of antigenic properties of macromolecules over 70 Myr". Nature. 252 (5478): 63–64. Bibcode:1974Natur.252...63D. doi:10.1038/252063a0. PMID 4139661. S2CID 4286786.
- ^ Jump up to: a b Westbrock P, van der Meide PH; van der Wey-Kloppers JS; et al. (1979). "Fossil macromolecules from cephalopod shells: Characterization, immunological response and diagenesis". Paleobiology. 5 (2): 151–167. doi:10.1017/S0094837300006448.
- ^ Jump up to: a b Nitecki MH (1982). Biochemical Aspects of Evolutionary Biology. Chicago: University of Chicago Press. pp. 29–91.
- ^ Jump up to: a b Gurley, L. R; J. G. Valdez; W. D. Spall; B. F Smith; D. D. Gillette (February 1991). "Proteins in the fossil bone of the dinosaur, seismosaurus". Journal of Protein Chemistry. 10 (1): 75–90. doi:10.1007/BF01024658. PMID 2054066. S2CID 9046093.
- ^ Jump up to: a b Kaiser, J. (7 October 2005). "VIROLOGY: Resurrected Influenza Virus Yields Secrets of Deadly 1918 Pandemic" (PDF). Science. 310 (5745): 28–29. doi:10.1126/science.310.5745.28. PMID 16210501. S2CID 26252589. Retrieved 9 April 2014.
- ^ Jump up to: a b Rockefeller University (1 March 2007). "Ancient Retrovirus Is Resurrected". Science Daily. Retrieved 9 April 2014.
- ^ Jump up to: a b Krause, Johannes; Fu, Qiaomei; Good, Jeffrey M.; Viola, Bence; Shunkov, Michael V.; Derevianko, Anatoli P.; Pääbo, Svante (24 March 2010). "The complete mitochondrial DNA genome of an unknown hominin from southern Siberia". Nature. 464 (7290): 894–897. Bibcode:2010Natur.464..894K. doi:10.1038/nature08976. PMID 20336068.
- ^ Jump up to: a b c Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PL, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Pääbo S (18 December 2013). "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature. 505 (7481): 43–49. Bibcode:2014Natur.505...43P. doi:10.1038/nature12886. PMC 4031459. PMID 24352235.
- ^ Jump up to: a b c Callaway, Ewen (4 December 2013). "Hominin DNA baffles experts". Nature. 504 (7478): 16–17. Bibcode:2013Natur.504...16C. doi:10.1038/504016a. PMID 24305130.
- ^ Jump up to: a b Dalton, Rex (15 May 2006). "Neanderthal DNA yields to genome foray". Nature. 441 (7091): 260–1. Bibcode:2006Natur.441..260D. doi:10.1038/441260b. PMID 16710377.
- ^ Jump up to: a b Zimmer, Carl (16 November 2015). "In a Tooth, DNA From Some Very Old Cousins, the Denisovans". New York Times. Retrieved 16 November 2015.
- ^ Jump up to: a b Sawyer, Susanna; Renaud, Gabriel; Viola, Bence; Hublin, Jean-Jacques; Gansauge, Marie-Theres; Shunkov, Michael V.; Derevianko, Anatoly P.; Prüfer, Kay; Kelso, Janet; Pääbo, Svante (11 November 2015). "Nuclear and mitochondrial DNA sequences from two Denisovan individuals". PNAS. 112 (51): 15696–700. Bibcode:2015PNAS..11215696S. doi:10.1073/pnas.1519905112. PMC 4697428. PMID 26630009.
- ^ Higuchi, Russell; Bowman, Barbara; Freiberger, Mary; Ryder, Oliver A.; Wilson, Allan C. (15 November 1984). "DNA sequences from the quagga, an extinct member of the horse family". Nature. 312 (5991): 282–284. Bibcode:1984Natur.312..282H. doi:10.1038/312282a0. PMID 6504142. S2CID 4313241.
- ^ Krause, Johannes; Fu, Qiaomei; Good, Jeffrey M.; Viola, Bence; Shunkov, Michael V.; Derevianko, Anatoli P.; Pääbo, Svante (24 March 2010). "The complete mitochondrial DNA genome of an unknown hominin from southern Siberia". Nature. 464 (7290): 894–897. Bibcode:2010Natur.464..894K. doi:10.1038/nature08976. PMID 20336068.
- ^ Jump up to: a b Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, Maricic T, Good JM, Marques-Bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M, Richards M, Talamo S, Shunkov MV, Derevianko AP, Hublin JJ, Kelso J, Slatkin M, Pääbo S (23 December 2010). "Genetic history of an archaic hominin group from Denisova Cave in Siberia". Nature. 468 (7327): 1053–1060. Bibcode:2010Natur.468.1053R. doi:10.1038/nature09710. PMC 4306417. PMID 21179161.
- ^ Pennisi, E. (16 May 2013). "More Genomes From Denisova Cave Show Mixing of Early Human Groups". Science. 340 (6134): 799. Bibcode:2013Sci...340..799P. doi:10.1126/science.340.6134.799. PMID 23687020.
- ^ Mounier, Aurélien; Marchal, François; Condemi, Silvana (March 2009). "Is Homo heidelbergensis a distinct species? New insight on the Mauer mandible". Journal of Human Evolution. 56 (3): 219–246. doi:10.1016/j.jhevol.2008.12.006. PMID 19249816.
- ^ Cartmill, Matt & Smith, Fred H. (2009). The Human Lineage. John Wiley & Sons. ISBN 978-0471214915. Retrieved 2013-04-21.
- ^ Jump up to: a b Meyer, Matthias; Fu, Qiaomei; Aximu-Petri, Ayinuer; Glocke, Isabelle; Nickel, Birgit; Arsuaga, Juan-Luis; Martínez, Ignacio; Gracia, Ana; de Castro, José María Bermúdez; Carbonell, Eudald; Pääbo, Svante (4 December 2013). "A mitochondrial genome sequence of a hominin from Sima de los Huesos" (PDF). Nature. 505 (7483): 403–406. Bibcode:2014Natur.505..403M. doi:10.1038/nature12788. PMID 24305051. S2CID 4456221. Archived from the original (PDF) on 12 February 2014. Retrieved 23 April 2014.
- ^ Zimmer, Carl. "Bringing Extinct Species Back To Life". National Geographic. Retrieved 26 March 2014.
- ^ Gray, Richard (31 Jan 2009). "Extinct ibex is resurrected by cloning". Telegraph. Archived from the original on 2 August 2009. Retrieved 9 April 2014.
- ^ Church, George. "Multiplex Automated Genomic Engineering (MAGE): A machine that speeds up evolution is revolutionizing genome design". Wyss Institute. Retrieved 26 March 2014.
- ^ Jump up to: a b The Editors (14 May 2013). "Why Efforts to Bring Extinct Species Back from the Dead Miss the Point". Scientific American. Retrieved 9 April 2014.
- ^ Pimm, S. L.; Russell, G. J.; Gittleman, J. L.; Brooks, T. M. (21 July 1995). "The Future of Biodiversity" (PDF). Science. 269 (5222): 347–350. Bibcode:1995Sci...269..347P. doi:10.1126/science.269.5222.347. PMID 17841251. S2CID 35154695. Retrieved 9 April 2014.
- ^ Hogan, Michael. "Causes of Extinction". The Encyclopedia of Earth. Retrieved 9 April 2014.
- ^ Church, George (20 August 2013). "De-Extinction Is a Good Idea". Scientific American. Retrieved 9 April 2014.
- ^ "Method: Principles". Amino Acid Geochronology Laboratory. Northern Arizona University. Archived from the original on 14 March 2012. Retrieved 26 March 2014.
- ^ Jump up to: a b c Poinar, H. N.; Hoss, M.; Bada, J. L.; Paabo, S. (10 May 1996). "Amino Acid Racemization and the Preservation of Ancient DNA". Science. 272 (5263): 864–866. Bibcode:1996Sci...272..864P. doi:10.1126/science.272.5263.864. hdl:2060/19980202342. PMID 8629020. S2CID 20626782.
- ^ Henn, B. M.; Gignoux, C. R.; Feldman, M. W.; Mountain, J. L. (6 October 2008). "Characterizing the Time Dependency of Human Mitochondrial DNA Mutation Rate Estimates". Molecular Biology and Evolution. 26 (1): 217–230. doi:10.1093/molbev/msn244. PMID 18984905.
- ^ "Mitochondrial DNA". DNA Analyst Training. NFSTC Science Serving Justice. Archived from the original on 1 February 2014. Retrieved 23 April 2014.
- ^ Lang, BF; Gray, MW; Burger, G (1999). "Mitochondrial genome evolution and the origin of eukaryotes". Annual Review of Genetics. 33: 351–97. doi:10.1146/annurev.genet.33.1.351. PMID 10690412.
- ^ Jump up to: a b Cano, RJ; Poinar, HN (Sep 1993). "Rapid isolation of DNA from fossil and museum specimens suitable for PCR". BioTechniques. 15 (3): 432–4, 436. PMID 8217155.
- ^ Jump up to: a b Tagliavia, Marcello; Massa, Bruno; Albanese, Ida; La Farina, Mario (29 March 2011). "DNA Extraction From Orthoptera Museum Specimens" (PDF). Analytical Letters. 44 (6): 1058–1062. doi:10.1080/00032719.2010.506939. hdl:10447/57711. S2CID 95380144.
- ^ Jump up to: a b c d e S. Blair Hodges; Mary H. Schweitzer. Molecular Paleontology (PDF). Encyclopedia of Paleontology. pp. 752–754.
- ^ Jump up to: a b Tuross, N (1994). "The biochemistry of ancient DNA in bone". Experientia. 50 (6): 50:530–535. doi:10.1007/bf01921721. PMID 7517371. S2CID 27896535.
- ^ Tuross, N; Stathoplos, L (1993). Ancient proteins in fossil bones. Methods in Enzymology. 224. pp. 121–129. doi:10.1016/0076-6879(93)24010-r. ISBN 9780121821258. PMID 8264383.
- Paleobiology
- Evolutionary biology
- Subfields of paleontology
- Ancient DNA (human)
- Fossils
