Virus classification
Virus classification is the process of naming viruses and placing them into a taxonomic system similar to the classification systems used for cellular organisms.
Viruses are classified by phenotypic characteristics, such as morphology, nucleic acid type, mode of replication, host organisms, and the type of disease they cause. The formal taxonomic classification of viruses is the responsibility of the International Committee on Taxonomy of Viruses (ICTV) system, although the Baltimore classification system can be used to place viruses into one of seven groups based on their manner of mRNA synthesis. Specific naming conventions and further classification guidelines are set out by the ICTV.
A catalogue of all the world's known viruses has been proposed and, in 2013, some preliminary efforts were underway.[1]
Virus species definition[]
Species form the basis for any biological classification system. Before 1982, it was thought that viruses could not be made to fit Ernst Mayr's reproductive concept of species, and so were not amenable to such treatment. In 1982, the ICTV started to define a species as "a cluster of strains" with unique identifying qualities. In 1991, the more specific principle that a virus species is a polythetic class of viruses that constitutes a replicating lineage and occupies a particular ecological niche was adopted.[2]
In July 2013, the ICTV definition of species changed to state: "A species is a monophyletic group of viruses whose properties can be distinguished from those of other species by multiple criteria."[3] These criteria include the structure of the capsid, the existence of an envelope, the gene expression program for its proteins, host range, pathogenicity, and most importantly genetic sequence similarity and phylogenetic relationship.[4]
The actual criteria used vary by the taxon, and can be inconsistent (arbitrary similarity thresholds) or unrelated to lineage (geography) at times.[5] The matter is, for many, not yet settled.[2]
ICTV classification[]
The International Committee on Taxonomy of Viruses began to devise and implement rules for the naming and classification of viruses early in the 1970s, an effort that continues to the present. The ICTV is the only body charged by the International Union of Microbiological Societies with the task of developing, refining, and maintaining a universal virus taxonomy.[6] The system shares many features with the classification system of cellular organisms, such as taxon structure. However, some differences exist, such as the universal use of italics for all taxonomic names, unlike in the International Code of Nomenclature for algae, fungi, and plants and International Code of Zoological Nomenclature.[7]
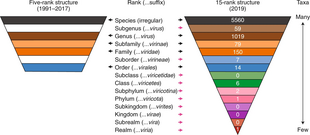
Viral classification starts at the level of realm and continues as follows, with the taxonomic suffixes in parentheses:[7]
- Realm (-viria)
- Subrealm (-vira)
- Kingdom (-virae)
- Subkingdom (-virites)
- Phylum (-viricota)
- Subphylum (-viricotina)
- Class (-viricetes)
- Subclass (-viricetidae)
- Order (-virales)
- Suborder (-virineae)
- Family (-viridae)
- Subfamily (-virinae)
- Genus (-virus)
- Subgenus (-virus)
- Species
- Subgenus (-virus)
- Genus (-virus)
- Subfamily (-virinae)
- Family (-viridae)
- Suborder (-virineae)
- Order (-virales)
- Subclass (-viricetidae)
- Class (-viricetes)
- Subphylum (-viricotina)
- Phylum (-viricota)
- Subkingdom (-virites)
- Kingdom (-virae)
- Subrealm (-vira)
Unlike the system of binomial nomenclature adopted in cellular species, there is currently no standardized form for virus species names. At present, the ICTV mandates that a species name must contain as few words as possible while remaining distinct, and must not only contain the word virus and the host name.[8] Species names often take the form of [Disease] virus, particularly for higher plants and animals. In 2019, the ICTV published a proposal to adopt a more formalized system of binomial nomenclature for virus species names, to be voted on in 2020.[9] However, some virologists later objected to the potential naming system change, arguing that the debate came while many in the field were preoccupied due to the COVID-19 pandemic.[10]
As of 2019, all levels of taxa except subrealm, subkingdom, and subclass are used. Four realms, one incertae sedis order, 24 incertae sedis families, and three incertae sedis genera are recognized:[11]
Realms: Duplodnaviria, Monodnaviria, Riboviria, and Varidnaviria
Incertae sedis order: Ligamenvirales
Incertae sedis families:
- Alphasatellitidae
- Ampullaviridae
- Anelloviridae
- Avsunviroidae
- Baculoviridae
- Bicaudaviridae
- Clavaviridae
- Finnlakeviridae
- Fuselloviridae
- Globuloviridae
- Guttaviridae
- Halspiviridae
- Hytrosaviridae
- Nimaviridae
- Nudiviridae
- Ovaliviridae
- Plasmaviridae
- Polydnaviridae
- Portogloboviridae
- Pospiviroidae
- Spiraviridae
- Thaspiviridae
- Tolecusatellitidae
- Tristromaviridae
Incertae sedis genera: Deltavirus, Dinodnavirus, Rhizidiovirus
Structure-based virus classification[]
It has been suggested that similarity in virion assembly and structure observed for certain viral groups infecting hosts from different domains of life (e.g., bacterial tectiviruses and eukaryotic adenoviruses or prokaryotic Caudovirales and eukaryotic herpesviruses) reflects an evolutionary relationship between these viruses.[12] Therefore, structural relationship between viruses has been suggested to be used as a basis for defining higher-level taxa – structure-based viral lineages – that could complement the ICTV classification scheme of 2010.[13]
The ICTV has gradually added many higher-level taxa using relationships in protein folds. All four realms defined in the 2019 release are defined by the presence of a protein of a certain structural family.[14]
Baltimore classification[]

Baltimore classification (first defined in 1971) is a classification system that places viruses into one of seven groups depending on a combination of their nucleic acid (DNA or RNA), strandedness (single-stranded or double-stranded), sense, and method of replication. Named after David Baltimore, a Nobel Prize-winning biologist, these groups are designated by Roman numerals. Other classifications are determined by the disease caused by the virus or its morphology, neither of which are satisfactory due to different viruses either causing the same disease or looking very similar. In addition, viral structures are often difficult to determine under the microscope. Classifying viruses according to their genome means that those in a given category will all behave in a similar fashion, offering some indication of how to proceed with further research. Viruses can be placed in one of the seven following groups:[15]
- I: dsDNA viruses (e.g. Adenoviruses, Herpesviruses, Poxviruses)
- II: ssDNA viruses (+ strand or "sense") DNA (e.g. Parvoviruses)
- III: dsRNA viruses (e.g. Reoviruses)
- IV: (+)ssRNA viruses (+ strand or sense) RNA (e.g. Coronaviruses, Picornaviruses, Togaviruses)
- V: (−)ssRNA viruses (− strand or antisense) RNA (e.g. Orthomyxoviruses, Rhabdoviruses)
- VI: ssRNA-RT viruses (+ strand or sense) RNA with DNA intermediate in life-cycle (e.g. Retroviruses)
- VII: dsDNA-RT viruses DNA with RNA intermediate in life-cycle (e.g. Hepadnaviruses)

DNA viruses[]
Viruses with a DNA genome, except for the DNA reverse transcribing viruses, are members of three of the four recognized viral realms: Duplodnaviria, Monodnaviria, and Varidnaviria. But the incertae sedis order Ligamenvirales, and many other incertae sedis families and genera, are also used to classify DNA viruses. The domains Duplodnaviria and Varidnaviria consist of double-stranded DNA viruses; other double-stranded DNA viruses are incertae sedis. The domain Monodnaviria consists of single-stranded DNA viruses that generally encode a HUH endonuclease; other single-stranded DNA viruses are incertae sedis.[11]
- Group I: viruses possess double-stranded DNA. Viruses that cause chickenpox and herpes are found here.
- Group II: viruses possess single-stranded DNA.
| Virus family | Examples (common names) | Virion naked/enveloped |
Capsid symmetry |
Nucleic acid type | Group |
|---|---|---|---|---|---|
| 1. Adenoviridae | Canine hepatitis virus, Some types of the common cold | Naked | Icosahedral | ds | I |
| 2. Papovaviridae | JC virus, HPV | Naked | Icosahedral | ds circular | I |
| 3. Parvoviridae | Human parvovirus B19, canine parvovirus | Naked | Icosahedral | ss | II |
| 4. Herpesviridae | Herpes simplex virus, varicella-zoster virus, cytomegalovirus, Epstein–Barr virus | Enveloped | Icosahedral | ds | I |
| 5. Poxviridae | Smallpox virus, cowpox, myxoma virus, monkeypox, vaccinia virus | Complex coats | Complex | ds | I |
| 6. Anelloviridae | Torque teno virus | Naked | Icosahedral | ss circular | II |
| 7. Pleolipoviridae | HHPV1, HRPV1 | Enveloped | ss/ds linear/circular | I/II |
RNA viruses[]
All viruses that have an RNA genome, and that encode an RNA-dependent RNA polymerase (RdRp), are members of the kingdom Orthornavirae, within the realm Riboviria.[16]
- Group III: viruses possess double-stranded RNA genomes, e.g. rotavirus.
- Group IV: viruses possess positive-sense single-stranded RNA genomes. Many well known viruses are found in this group, including the picornaviruses (which is a family of viruses that includes well-known viruses like Hepatitis A virus, enteroviruses, rhinoviruses, poliovirus, and foot-and-mouth virus), SARS virus, hepatitis C virus, yellow fever virus, and rubella virus.
- Group V: viruses possess negative-sense single-stranded RNA genomes. Ebola and Marburg viruses are well known members of this group, along with influenza virus, measles, mumps and rabies.
| Virus Family | Examples (common names) | Capsid naked/enveloped |
Capsid Symmetry |
Nucleic acid type | Group |
|---|---|---|---|---|---|
| 1. Reoviridae | Reovirus, rotavirus | Naked | Icosahedral | ds | III |
| 2. Picornaviridae | Enterovirus, rhinovirus, hepatovirus, cardiovirus, aphthovirus, poliovirus, parechovirus, erbovirus, kobuvirus, teschovirus, coxsackie | Naked | Icosahedral | ss | IV |
| 3. Caliciviridae | Norwalk virus | Naked | Icosahedral | ss | IV |
| 4. Togaviridae | Eastern equine encephalitis | Enveloped | Icosahedral | ss | IV |
| 5. Arenaviridae | Lymphocytic choriomeningitis virus, Lassa fever | Enveloped | Complex | ss(-) | V |
| 6. Flaviviridae | Dengue virus, hepatitis C virus, yellow fever virus, Zika virus | Enveloped | Icosahedral | ss | IV |
| 7. Orthomyxoviridae | Influenzavirus A, influenzavirus B, influenzavirus C, isavirus, thogotovirus | Enveloped | Helical | ss(-) | V |
| 8. Paramyxoviridae | Measles virus, mumps virus, respiratory syncytial virus, Rinderpest virus, canine distemper virus | Enveloped | Helical | ss(-) | V |
| 9. Bunyaviridae | California encephalitis virus, Sin nombre virus | Enveloped | Helical | ss(-) | V |
| 10. Rhabdoviridae | Rabies virus, Vesicular stomatitis | Enveloped | Helical | ss(-) | V |
| 11. Filoviridae | Ebola virus, Marburg virus | Enveloped | Helical | ss(-) | V |
| 12. Coronaviridae | Human coronavirus 229E, Human coronavirus NL63, Human coronavirus OC43, Human coronavirus HKU1, Middle East respiratory syndrome-related coronavirus, Severe acute respiratory syndrome coronavirus, and Severe acute respiratory syndrome coronavirus 2 | Enveloped | Helical | ss | IV |
| 13. Astroviridae | Astrovirus | Naked | Icosahedral | ss | IV |
| 14. Bornaviridae | Borna disease virus | Enveloped | Helical | ss(-) | V |
| 15. Arteriviridae | Arterivirus, equine arteritis virus | Enveloped | Icosahedral | ss | IV |
| 16. Hepeviridae | Hepatitis E virus | Naked | Icosahedral | ss | IV |
Reverse transcribing viruses[]
All viruses that encode a reverse transcriptase (also known as RT or RNA-dependent DNA polymerase) are members of the class Revtraviricetes, within the phylum Arterviricota, kingdom Pararnavirae, and realm Riboviria. The class Blubervirales contains the single family Hepadnaviridae of DNA RT (reverse transcribing) viruses; all other RT viruses are members of the class Ortervirales.[17]
- Group VI: viruses possess single-stranded RNA viruses that replicate through a DNA intermediate. The retroviruses are included in this group, of which HIV is a member.
- Group VII: viruses possess double-stranded DNA genomes and replicate using reverse transcriptase. The hepatitis B virus can be found in this group.
| Virus Family | Examples (common names) | Capsid naked/enveloped |
Capsid Symmetry |
Nucleic acid type | Group |
|---|---|---|---|---|---|
| 1. Retroviridae | HIV | Enveloped | dimer RNA | VI | |
| 2. Caulimoviridae | Caulimovirus, Cacao swollen-shoot virus (CSSV) | Naked | VII | ||
| 3. Hepadnaviridae | Hepatitis B virus | Enveloped | Icosahedral | circular, partially ds | VII |
Historical systems[]
Holmes classification[]
(1948) used a Linnaean taxonomy with binomial nomenclature to classify viruses into 3 groups under one order, . They are placed as follows:
- Group I: Phaginae (attacks bacteria)
- Group II: Phytophaginae (attacks plants)
- Group III: Zoophaginae (attacks animals)
The system was not accepted by others due to its neglect of morphological similarities.[18]
Subviral agents[]
The following infectious agents are smaller than viruses and have only some of their properties.[19][20] Since 2015, the ICTV has allowed them to be classified in a similar way as viruses are.[21]
Viroids and virus-dependent agents[]
Viroids[]
- Family Avsunviroidae[22]
- Genus Avsunviroid; type species: Avocado sunblotch viroid
- Genus Pelamoviroid; type species: Peach latent mosaic viroid
- Genus Elaviroid; type species:
- Family Pospiviroidae[23]
- Genus Pospiviroid; type species: Potato spindle tuber viroid
- Genus Hostuviroid; type species: Hop stunt viroid
- Genus Cocadviroid; type species: Coconut cadang-cadang viroid
- Genus Apscaviroid; type species:
- Genus Coleviroid; type species:
Satellites[]
Satellites depend on co-infection of a host cell with a helper virus for productive multiplication. Their nucleic acids have substantially distinct nucleotide sequences from either their helper virus or host. When a satellite subviral agent encodes the coat protein in which it is encapsulated, it is then called a satellite virus.
Satellite-like nucleic acids resemble satellite nucleic acids, in that they replicate with the aid of helper viruses. However they differ in that they can encode functions that can contribute to the success of their helper viruses; while they are sometimes considered to be genomic elements of their helper viruses, they are not always found within their helper viruses.[19]
- Satellite viruses[24]
- Single-stranded RNA satellite viruses
- (unnamed family)
- Aumaivirus – Maize white line mosaic satellite virus
- Papanivirus – Panicum mosaic satellite virus
- Virtovirus – Tobacco mosaic satellite virus
- Albetovirus – Tobacco necrosis satellite virus
- Family
- – (extra small virus)
- (unnamed genus) –
- (unnamed genus) –
- (unnamed family)
- Double-stranded DNA satellite viruses
- Family Lavidaviridae – Virophages
- Single-stranded DNA satellite viruses
- Genus Dependoparvovirus – Adeno-associated virus group
- Single-stranded RNA satellite viruses
- Satellite nucleic acids
- Single-stranded satellite DNAs
- Family Alphasatellitidae (encoding a replication initiator protein)
- Family Tolecusatellitidae (encoding a pathogenicity determinant βC1)
- Double-stranded satellite RNAs
- Single-stranded satellite RNAs
- Subgroup 1: Large satellite RNAs
- Subgroup 2: Small linear satellite RNAs
- Subgroup 3: Circular satellite RNAs (virusoids)
- Genus Deltavirus
- Polerovirus-associated RNAs
- Satellite-like RNA
- Satellite-like DNA
- Single-stranded satellite DNAs
Defective interfering particles[]
Defective interfering particles are defective viruses that have lost their ability to replicate except in the presence of a helper virus, which is normally the parental virus. They can also interfere with the helper virus.
- Defective interfering particles (RNA)
- Defective interfering particles (DNA)
See also[]
- Glossary of scientific naming
- Binomial nomenclature
- Biological classification
- International Committee on Taxonomy of Viruses
- List of genera of viruses
- List of virus species
- Nomenclature codes
- Prion
- Taxonomy
- Trinomial nomenclature
- Virology
Notes[]
- ^ Zimmer C (5 September 2013). "A Catalog for All the World's Viruses?". New York Times. Retrieved 6 September 2013.
- ^ Jump up to: a b Alimpiev, Egor (March 15, 2019). Rethinking the Virus Species Concept (PDF).
- ^ Adams MJ, Lefkowitz EJ, King AM, Carstens EB (December 2013). "Recently agreed changes to the International Code of Virus Classification and Nomenclature". Archives of Virology. 158 (12): 2633–9. doi:10.1007/s00705-013-1749-9. PMID 23836393.
- ^ "International Committee on Taxonomy of Viruses (ICTV)". International Committee on Taxonomy of Viruses (ICTV). Retrieved 2021-06-10.
- ^ Peterson, A Townsend (23 July 2014). "Defining viral species: making taxonomy useful". Virology Journal. 11 (1): 131. doi:10.1186/1743-422X-11-131. PMC 4222810. PMID 25055940.
- ^ Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB (January 2018). "Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV)". Nucleic Acids Research. 46 (D1): D708–D717. doi:10.1093/nar/gkx932. PMC 5753373. PMID 29040670.
- ^ Jump up to: a b "ICTV Code". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 26 April 2020.
- ^ "The International Code of Virus Classification and Nomenclature". ICTV. International Committee on Taxonomy of Viruses. Retrieved 2 September 2020.
- ^ Siddell, Stuart; Walker, Peter; Lefkowitz, Elliot; Mushegian, Arcady; Dutilh, Bas; Harrach, Balázs; Harrison, Robert; Junglen, Sandra; Knowles, Nick; Kropinski, Andrew; Krupovic, Mart; Kuhn, Jens; Nibert, Max; Rubino, Luisa; Sabanadzovic, Sead; Simmonds, Peter; Varsani, Arvind; Zerbini, Francisco; Davison, Andrew (3 December 2019). "Binomial nomenclature for virus species: a consultation". Arch Virol. 165 (2): 519–525. doi:10.1007/s00705-019-04477-6. PMC 7026202. PMID 31797129.
- ^ Mallapaty, Smriti (30 July 2020). "Should virus-naming rules change during a pandemic? The question divides virologists". Nature. 584 (7819): 19–20. Bibcode:2020Natur.584...19M. doi:10.1038/d41586-020-02243-2. PMID 32733098.
- ^ Jump up to: a b "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 26 April 2020.
- ^ Bamford DH (May 2003). "Do viruses form lineages across different domains of life?". Research in Microbiology. 154 (4): 231–6. doi:10.1016/S0923-2508(03)00065-2. PMID 12798226.
- ^ Krupovič M, Bamford DH (December 2010). "Order to the viral universe". Journal of Virology. 84 (24): 12476–9. doi:10.1128/JVI.01489-10. PMC 3004316. PMID 20926569.
- ^ "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 26 April 2020.
- ^ "Baltimore Classification of Viruses" (Website.) Molecular Biology Web Book - http://web-books.com/. Retrieved on 2008-08-18.
- ^ "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 25 April 2020.
- ^ Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH. "Proposal: Create a megataxonomic framework, filling all principal taxonomic ranks, for realm Riboviria". International Committee on Taxonomy of Viruses (ICTV). Retrieved 2020-05-21.CS1 maint: multiple names: authors list (link)
- ^ Kuhn, Jens H. (2020). "Virus Taxonomy". Reference Module in Life Sciences. Encyclopedia of Virology. pp. 28–37. doi:10.1016/B978-0-12-809633-8.21231-4. ISBN 978-0-12-809633-8. PMC 7157452.
- ^ STRAUSS, JAMES H.; STRAUSS, ELLEN G. (2008). "Subviral Agents". Viruses and Human Disease. Elsevier. pp. 345–368. doi:10.1016/b978-0-12-373741-0.50012-x. ISBN 978-0-12-373741-0. S2CID 80872659.
- ^ TaxoProp 2015.002aG
- ^ "80.002 Avsunviroidae - ICTVdB Index of Viruses." (Website.) U.S. National Institutes of Health website. Retrieved on 2007-09-27.
- ^ "80.001 Popsiviroidae - ICTVdB Index of Viruses." (Website.) U.S. National Institutes of Health website. Retrieved on 2007-09-27.
- ^ Krupovic, Mart; Kuhn, Jens H.; Fischer, Matthias G. (7 October 2015). "A classification system for virophages and satellite viruses". Archives of Virology. 161 (1): 233–247. doi:10.1007/s00705-015-2622-9. PMID 26446887.
External links[]
| Wikispecies has information related to Virus. |
- Virus taxonomy
- Biological classification

