Glossary of evolutionary biology
This glossary of evolutionary biology is a list of definitions of terms and concepts used in the study of evolutionary biology, population biology, speciation, and phylogenetics, as well as sub-disciplines and related fields. For additional terms from related glossaries, see Glossary of genetics, Glossary of ecology, and Glossary of biology.
A[]
- adaptation
- 1. The dynamic evolutionary process by which biological organisms develop characteristics that allow them to survive and reproduce within their environments.
- 2. The state or condition reached by a population during that process.
- 3. Any characteristic or phenotypic trait with a functional role in an individual organism and which has evolved and is maintained through natural selection.
- adaptationism
- The Darwinian view that many or most physiological and behavioral traits of organisms are adaptations that have evolved for specific functions or for specific reasons (as opposed to being byproducts of the evolution of other traits, consequences of biological constraints, or the result of random variation).
- adaptive radiation
- The simultaneous or near-simultaneous evolutionary divergence of multiple members of a single phylogenetic lineage into a variety of different forms with different adaptations, especially a diversification in the use of resources or habitats.[1]
- agamospecies
- A species that does not reproduce sexually but rather by cloning.[2] Agamospecies are sometimes represented by species complexes that contain some diploid individuals and other apomictic forms—in particular, plant species that can reproduce via agamospermy.[3]
- allele frequency
- allochronic isolation
- The isolation of two populations of a species due to a change in breeding periods. This isolation acts as a precursor to allochronic speciation, a type of speciation which results when two populations of a species become isolated due to differences in reproductive timing. An example is the periodical 13- and 17-year Magicicada species.[3]
- allo-parapatric speciation
- A mode of speciation where divergence occurs in allopatry and is completed upon secondary contact of the populations--effectively a form of reinforcement.[4][3]
- allometry
- The comparative study of the relationship between the size of an organism's body (or of a specific organ, e.g. the brain) and various other biological characteristics, such as body shape, anatomy, physiology, or behavior.
- allopatric speciation
- A mode of speciation where the evolution of reproductive isolation is caused by the geographic separation of two or more populations of a single species.[5]
- allopatric taxa
- Specific species that are allopatrically distributed.
- allopatry
- The phenomenon by which two or more populations of a single species exist in geographic isolation from one another.
- allopolyploid
- A polyploid cell or organism in which the several sets of chromosomes originate from more than one species, as in an intraspecific hybrid.[1]
- allo-sympatric speciation
- A mode of speciation where divergence occurs in allopatry and is completed upon secondary contact of the populations–effectively a form of reinforcement.[6][3]
- altruism
- anagenesis
- Evolutionary change that occurs within a species lineage as opposed to lineage splitting (cladogenesis).[7]
- ancestral trait
- For a given clade, any trait or feature (e.g. a specific phenotype) that appears in the clade's common ancestor; the same trait may also appear in some or all of the lineal descendants included within the clade, indicating that it has undergone little or no significant change during the clade's evolutionary history and thus retained its "primitive" condition. Some but not all subgroups within the clade may contain derived traits, in which the ancestral trait has changed significantly over evolutionary time such that the original ancestral condition no longer exists. Both terms are relative: an ancestral trait for one clade may be a derived trait for a different clade. The term "ancestral trait" is often used interchangeably with the more technical term plesiomorphy.
- apomorphy
- A derived character state; i.e. the state or condition of a particular trait or feature (e.g. a specific phenotype) that is distinct from and derivative of an ancestral character by virtue of its modification over time in one or more lineal descendants of a given clade. Apomorphies are often viewed as evolutionary "innovations" which set the taxa in which they appear apart from the clade's common ancestor, as well as from other clades; shared apomorphies are used to construct and define clades. The term is relative; a trait considered an apomorphy in one clade may not be considered an apomorphy in a different clade. Contrast plesiomorphy.
- area cladogram
- asexual reproduction
- assortative mating
- A mating system in which individuals with similar phenotypes mate with each other more frequently than would be expected in a completely random mating system. Assortative mating usually has the effect of increasing genetic relatedness between members of the mating population. Contrast disassortative mating.
- autapomorphy
- autoallopolyploid
- autopolyploid

In allopatric speciation, a population becomes separated by a geographic barrier and reproductive isolation results in two separate species

An apomorphy is a derived trait present in one or more members of a clade but not the common ancestor; a plesiomorphy is an ancestral trait present in the common ancestor of the clade and possibly some or all of its descendants.
B[]
- barrier
- Bateson–Dobzhansky–Muller model
- An evolutionary model of the genetic incompatibility that occurs as a result of negative epistatic interactions between two or more genes or alleles with different evolutionary histories, which may meet when distinct populations hybridize. The incompatible genes or alleles themselves, referred to as Dobzhansky–Muller incompatibilities, may be the result of random or neutral mutations, or they may be specific adaptations driven by natural selection. By preventing populations from successfully interbreeding, these incompatibilities can reinforce reproductive isolation and thereby increase the chance of speciation.
- behavioral isolation
- biogeography
- The scientific study of the spatial distributions of biological organisms, populations, and species. It includes the study of both extinct and extant organisms.[8]
- biological constraints
- biological species concept
- bottleneck
- See population bottleneck.
C[]
- centrifugal speciation
- A variation of peripatric speciation where speciation occurs by geographic isolation, but reproductive isolation evolves in the larger population instead of the peripherally isolated population.[9]
- chromosomal speciation
- chronospecies
- clade
- A phylogenetic grouping of organisms that consists of a single common ancestor and all of its lineal descendants, and which by definition is monophyletic. The common ancestor may be an individual organism, a population, a species, or any other taxon; any and all members of a clade may be extant or extinct. Clades can be visualized with cladograms and are the basis of cladistics.
- cladistics
- An approach to biological classification in which organisms are grouped in clades defined by shared ancestry; hypothesized relationships between organisms are typically based on shared derived characters which can be traced to the most recent common ancestor and are not present in more distant ancestors or unrelated groups.
- cladogenesis
- The splitting of a single species lineage within a phylogeny into multiple lineages.[7]
- cladogram
- cline
- A measurable spatial gradient in a single biological character or trait of a species or population across its geographic range. The nature of a cline may be genotypic (e.g. variation in allele frequency) or phenotypic (e.g. variation in body size or pigmentation), and may show smooth, continuous gradation or abrupt changes between different geographic regions.
- cluster analysis
- clustering
- character displacement
- co-operation
- coevolution
- The process by which two or more distinct populations, species, or other groups of organisms, or two or more distinct traits within a species, reciprocally affect each other's evolution through natural selection. Each party in a coevolutionary relationship exerts selective pressures upon the other, leading to the evolution of separate traits in each party.
- cohesion species concept
- colonization
- The spread of a population to a new area.
- common ancestor
- An organism or taxon (e.g. a species) which is hypothesized to be the lineal progenitor of two or more organisms or taxa which exist at a later point in evolutionary time. The concept of common descent is fundamental to the study of evolution, phylogenetics, and cladistics; for instance, all clades, by definition, are rooted in a common ancestor. See also most recent common ancestor.
- competitive gametic isolation
- congruent clines
- convergent evolution
- copulatory behavioral isolation
- coupling
- court jester hypothesis
- cospeciation
- A type of speciation in which more than two species speciate concurrently due to their ecological associations (e.g. host-parasite interactions).[10]
- crown group
- cryptic species
- cytoplasmic isolation
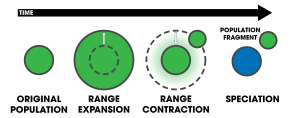
In centrifugal speciation, the range of an original population (green) expands and then contracts, leaving an isolated fragment population behind. In the absence of interbreeding, the central population (changed to blue) evolves reproductive isolation over time.
D[]
- Darwinism
- The understanding of biological evolution as developed by the English naturalist Charles Darwin and others, which states that all biological organisms arise and develop through the natural selection of small, inherited variations that increase the individual's ability to compete, survive, and reproduce. Colloquially, the term is sometimes used to refer more broadly to modern evolutionary theory as a whole, though in scientific circles distinctions are usually made between Darwin's ideas and later additions to evolutionary biology.
- de-extinction
- derived trait
- For a given clade, any trait or feature (e.g. a specific phenotype) that is present within one or more subgroups of the clade but not in the clade's common ancestor. Derived traits show significant differences from the original "primitive" condition of an ancestral trait found in the common ancestor, implying that the trait has undergone extensive adaptation during the clade's evolutionary history to reach its derivative condition. Both terms are relative: a derived trait for one clade may be an ancestral trait for a different clade. The term "derived trait" is often used interchangeably with the more technical term apomorphy.
- developmental biology
- directional selection
- A mode of natural selection in which an extreme trait or phenotype is favored over other phenotypes, causing allele frequencies to shift over time in the direction of that trait or phenotype. This shift can occur whether or not the alleles governing the extreme phenotype are dominant or recessive.
- directional speciation
- disassortative mating
- A mating system in which individuals with dissimilar phenotypes mate with each other more frequently than would be expected in a completely random mating system. Disassortative mating usually has the effect of decreasing genetic relatedness between members of the mating population. Contrast assortative mating.
- dispersal
- disruptive selection
- A mode of natural selection in which the extreme values of a trait or phenotype within a breeding population are favored over intermediate values, causing allele frequencies to shift over time away from the intermediate. This causes the variance in the trait to increase and results in the population dividing into two distinct groups, each with trait values at one end of the trait's distribution curve.
- divergence-with-gene-flow
- divergent evolution
- The process by which any phenotypic or genotypic distinction emerges between two different populations or evolutionary lineages. Divergence may occur by any of a variety of mechanisms but is often especially noticeable after the two lineages have been reproductively isolated for many generations.[7]
- diversification
- Dobzhansky–Muller model
- See Bateson–Dobzhansky–Muller model.
E[]
- ecogeographic isolation
- ecological allopatry
- ecological character displacement
- ecological isolation
- ecological niche
- ecological speciation
- A type of speciation in which reproductive isolation is caused by the interaction of individuals of a species with their environment.[11]
- ecological species concept
- endemism
- The ecological state of a species being unique to a single geographic location, such as an island, nation, country, or any other clearly defined area, or to a single habitat type.
- environmental gradient
- error catastrophe
- The extinction of a population of organisms (insofar as the population can be defined by one or more identifiable characteristics) as a result of the excessive accumulation of genetic mutations, such that the population loses self-identity because all of its mutated descendants lack the identifiable characteristics.
- ethological isolation
- ethological pollinator isolation
- evolution
- The phenomenon by which the heritable characteristics of biological populations change over successive generations. Evolution occurs when processes such as natural selection and genetic drift act on the variation in characteristics that exists between members of a population, resulting in certain characteristics becoming more or less common within the population.
- evolutionary arms race
- The positive feedback mechanism operating between competing sets of co-evolving genes, traits, species, or other taxa which evolve specific adaptations and counter-adaptations due to each other's presence, which may be seen as analogous with an "arms race".
- evolutionary biology
- The discipline of biology that studies the evolution of biological organisms and the processes by which it operates, including natural selection, adaptation, common descent, and speciation. A core element of the modern synthesis, evolutionary biology integrates concepts from genetics, systematics, ecology, paleontology, developmental biology, and numerous other fields.
- evolutionary landscape
- evolutionary lineage
- The line of descent of a species.[7]
- evolutionary species concept
- exaptation
- extant
- Currently living or existing; still in existence and not extinct. The term is generally used to refer to the present-day state of existence of a particular taxon (such as a family, genus, species, etc.).
- extended evolutionary synthesis
- extinction
- extrinsic hybrid inviability
- extrinsic postzygotic isolation
F[]
- fitness The reproductive success, or propensity to produce offspring, during the lifetime of an individual
- fixation
- The process by which a single allele for a particular gene with multiple alleles increases in frequency in a given population such that it becomes permanently established as the only allele at that locus within the population's gene pool. How long fixation takes depends on selection pressures and chance fluctuations in allele frequencies.
- floral isolation
- flowering asynchrony
- founder effect
- The loss of genetic variation that occurs when a new, physically isolated population is established by a very small number of individuals who have migrated from a larger population and are not fully representative of the larger population's genetic diversity. As a result, the new population is often distinctively different, both genotypically and phenotypically, from the parent population. Besides migration, population bottlenecks can also result in a type of founder effect; extreme founder effects can lead to speciation.
- founder event
- founder-flush-crash
- founder takes all
- A hypothesis that describes the evolutionary advantages of the first-arriving lineages in a new ecosystem.[12] An example could be when a species becomes reproductively isolated on an island, as in peripatric speciation.
- fugitive species
- A species that occupies temporary environments or habitats (either because its members frequently migrate or because its environments frequently change) and so does not persist for many generations at any one site.[1]
G[]
- gametic isolation
- gene
- Any segment or set of segments of a nucleic acid molecule that contains the information necessary to produce a functional RNA transcript in a controlled manner. Genes are often considered the fundamental units of heredity and are typically encoded in DNA. A particular gene can have multiple different versions, or alleles, and a single gene may influence many different phenotypes.
- gene flow
- The transfer of genetic variation from one population to another.
- gene pool
- The sum of all of the various alleles shared by the members of a single population.
- genealogical species concept
- generation
- The average period during which an individual is born and survives until reproduction. Also used in reference to a group of individuals in which this period overlaps.
- genetic bottleneck
- See population bottleneck.
- genetic distance
- A measure of the genetic divergence between species, populations within a species, or individuals, used especially in phylogenetics to express either the time elapsed since the existence of a common ancestor or the degree of differentiation in the DNA sequences comprising the genomes of each population or individual.
- genetic drift
- A change in the frequency with which an existing allele occurs in a population due to random variation in the distribution of alleles from one generation to the next. It is often interpreted as the role that random chance plays in determining whether a given allele becomes more or less common with each generation, irrespective of the influence of natural selection. Genetic drift may cause certain alleles, even otherwise advantageous ones, to disappear completely from the gene pool, thereby reducing genetic variation, or it may cause initially rare alleles, even neutral or deleterious ones, to become much more frequent or even fixed.
- genetic erosion
- genetic load
- Any reduction in the mean fitness of a population owing to the existence of one or more genotypes with lower fitness than that of the most fit genotype.[1]
- genetic variation
- The genetic differences both within and between populations, species, or other groups of organisms. It is often visualized as the variety of different alleles in the gene pools of different populations.
- genic selection
- A type of natural selection that occurs at the level of individual genes or alleles, in which the frequency of an allele within a breeding population is determined by its fitness averaged over the variety of genotypes in which it occurs; the differential propagation of different alleles within a population as a consequence of properties borne by the alleles themselves, rather than by the genotypes in which they are found.[1]
- genic speciation
- genotype
- genotypic cluster species
- geographic speciation
- grade
- gradualism
- Continuous evolutionary change within a species lineage.[7] See also phyletic gradualism.
- green-beard effect

Gene flow is the transfer of alleles from one population to another population through the migration of individual organisms between the populations
H[]
- habitat isolation
- heredity
- The passing on of phenotypic traits from parents to their offspring through reproduction. Offspring are said to inherit the genetic information of their parents.
- heteropatric speciation
- Haldane's rule
- A rule formulated by J.B.S. Haldane which states that if one sex of the hybrid offspring resulting from a cross between two incipient species is inviable or sterile, that sex is more likely to be the heterogametic sex (i.e. the one with two different sex chromosomes).[13]
- Hardy–Weinberg principle
- A principle of population genetics which states that allele and genotype frequencies of a population will remain constant from generation to generation in the absence of other evolutionary influences. In the simplest case of a randomly mating population of diploid organisms possessing a single locus with two alleles, A and a, with frequencies f(A) = p and f(a) = q, respectively, the expected genotype frequencies are f(AA) = p2 for AA homozygotes, f(aa) = q2 for aa homozygotes, and f(Aa) = 2pq for heterozygotes. In the absence of evolutionary forces such as natural selection, mutation, assortative mating, gene flow, and genetic drift, p and q will remain constant between generations, such that the population is said to be in Hardy–Weinberg equilibrium with respect to the locus in question.
- homology
- A similarity between a pair of structures, traits, or DNA sequences in different taxa that is due to shared ancestry.
- homoplasy
- homoploid recombinational speciation
- host race
- host-specific parasite
- host-specific species
- hybrid
- The offspring that results from combining the qualities of two organisms of different genera, species, breeds, or varieties through sexual reproduction. Hybrids may occur naturally or artificially, as during selective breeding of domesticated animals and plants. Reproductive barriers typically prevent hybridization between distantly related organisms, or at least ensure that hybrid offspring are sterile, but fertile hybrids may result in speciation.
- hybrid breakdown
- hybrid incompatibility
- hybrid inviability
- hybrid speciation
- hybrid sterility
- hybrid swarm
- hybrid zone
- A geographic area in which the ranges of two interbreeding species or populations overlap, allowing them to cross-fertilize and generate hybrid offspring. The formation of a hybrid zone is one of the four outcomes of secondary contact between divergent genetic lineages.
- hybridization
- The process by which a hybrid organism is produced from two parents of different genera, species, breeds, or varieties.
- hypermorphosis
- The exaggeration of one or more phenotypic features of a descendant organism compared to those of its ancestors due to an increase in the duration of ontogenetic development over evolutionary history.[1]
I[]
- identical ancestors point
- identical by descent (IBD)
- (of a gene or allele) Traceable back through an arbitrary number of generations without mutation to a common ancestor of the group of descendant organisms that carries the gene or allele.[1] A gene or allele present in a group of descendant organisms is said to be identical by descent to a gene or allele in a common ancestor of the group if both sequences are identical, indicating that the sequence has been passed down unmodified from the common ancestor to its descendants.
- inbreeding depression
- inclusive fitness
- The number of offspring equivalents that an individual organism rears, rescues, or otherwise supports through its behavior, regardless of whether or not the individual is actually a biological parent of the offspring equivalents. Inclusive fitness is one of two metrics of evolutionary success as defined by W.D. Hamilton in 1964, the other being personal fitness.
- incomplete speciation
- incipient species
- Any population that is in an early stage of speciation.
- interbreeding
- intrinsic postzygotic isolation
- introgression
- inviability
- isolating mechanism
- isolation
- isolation by distance
- iterative evolution
- The repeated evolution of similar phenotypic characteristics or traits in different organisms at different times during the evolutionary history of a clade,[1] a phenomenon which can result in the seeming de-extinction of an organism previously considered extinct.
- iteroparity
- A reproductive strategy characterized by multiple reproductive cycles during an individual organism's lifetime. Organisms that use such a strategy are said to be iteroparous. Iteroparity is usually contrasted with semelparity.
J[]
- Jordan's Law
K[]
- k-selection
- Kaneshiro model
- A model of peripatric speciation developed by Kenneth Y. Kanneshiro where a sexual species experiences a population bottleneck—that is, when the genetic variation is reduced due to small population size—mating discrimination among females may be altered by the decrease in courtship behaviors or displays of males. This allows sexual selection to give rise to novel sexual traits in the new population.[14]
- kin selection
- A form of genic selection by which alleles differ in their rates of propagation by influencing the survival or reproductive success of individuals who carry the same alleles by common descent (their kin).[1]
- koinophilia
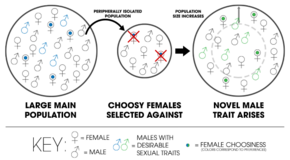
In the Kaneshiro model of peripatric speciation, a sample of a larger population results in an isolated population with less males containing attractive traits. Over time, choosy females are selected against as the population increases. Sexual selection drives new traits to arise (green), thereby reproductively isolating the new population from the old one (blue).
L[]
- last universal common ancestor (LUCA)
- The most recent population of organisms from which all extant organisms on Earth share a common descent; i.e. the most recent common ancestor of all organisms now living. LUCA is not thought to have been the earliest life on Earth, but rather the only organism of its time to still have living descendants. Its existence is not known from any specific fossil record but is inferred from phylogenetic comparisons of modern organisms, all of which are its descendants.
- lineage
- lineage-splitting
- When gene flow between two populations is completely eliminated.[7]
M[]
- macroevolution
- Evolutionary change as it occurs at a relatively large scale, at or above the level of species, as opposed to microevolution, which occurs at a smaller scale. Macroevolution is often thought of as the compounded effects of microevolution.
- maternal effect
- Any nongenetic effect of the mother on the phenotype of her offspring, owing to factors such as cytoplasmic inheritance, transmission of congenital disease, and the sharing of nutritional conditions.[1]
- mating system
- mating system isolation
- maximum parsimony
- See parsimony.
- mechanical isolation
- mechanical pollinator isolation
- meristic trait
- A discretely varying, countable trait, e.g. number of digits.[1]
- microallopatric
- Allopatric speciation occurring on a small geographic scale.[15]
- microevolution
- Evolutionary change as it occurs at a relatively small scale, typically within a particular species or population, as opposed to macroevolution, which occurs at a larger scale. Because of the convenience of observing and modeling small-scale changes in allele frequencies within discrete populations, the principles of population genetics are often conceptualized at microevolutionary scales.
- microspecies
- migration
- mimicry
- mitochondrial Eve
- modern synthesis
- modes of speciation
- A classification scheme of speciation processes based on the level of gene flow between two populations.[16] The traditional terms for the three modes—allopatric, parapatric, and sympatric—are based on the spatial distributions of a species population.[17][16]
- monophyly
- morphological species concept
- mosaic
- mosaic evolution
- mosaic hybrid zone
- A zone in which two speciating lineages occur together in a patchy distribution–either by chance, random colonization, or low hybrid fitness.[16]
- mosaic sympatry
- A case of sympatry in which two populations overlapping in geographic distribution exhibit habitat specializations.[16]
- most recent common ancestor (MRCA)
- Muller's ratchet
- multifurcation
- See polytomy.
- mutational meltdown
- mutationism
N[]
- natural selection
- network evolution
- See reticulate evolution.
- niche
- See ecological niche.
- niche adaptation
- niche preference
- noncompetitive gametic isolation
- nongenetic barrier
- non-geographic speciation
- norm of reaction
- See reaction norm.
O[]
- offspring
- ontogeny
- The origination and biological development of an organism within its own lifetime, as opposed to phylogeny, which refers to the evolutionary history of the organism's ancestors. In sexually reproducing organisms, ontogeny is the study of the development of an organism from the time of fertilization to the organism's reproductively mature form; the term may also be used to refer to the study of an organism's entire lifespan.
- operational taxonomic unit (OTU)
- orthogenesis
- outgroup
P[]
- paleopolyploidy
- para-allopatric speciation
- A mode of speciation in which divergence begins in parapatry but is completed in allopatry.[3]
- parallel evolution
- The independent evolution of similar or identical derived traits or characters in related lineages, thought usually to be based on similar modifications of common developmental pathways.[1] Contrast convergent evolution.
- parapatric speciation
- paraphyly
- parsimony
- The principle of accounting for empirical observations by whichever hypothesis requires the fewest or the simplest assumptions for which there is limited or no evidence. In biological systematics, maximum parsimony is an optimality criterion which invokes a minimum of evolutionary changes to infer phylogenetic relationships; i.e. the phylogenetic tree that minimizes the total number of character-state changes is to be preferred.[1]
- parthenogenesis
- A type of asexual reproduction in which the growth and development of embryos occurs without fertilization. In animals which reproduce by parthenogenesis, an unfertilized gamete of the female parent is capable of developing into an adult without any contribution from a male parent, resulting in offspring possessing only the mother's genetic material (the exact proportion of which depends on the parthenogenetic mechanism, of which there are numerous varieties). Some species reproduce exclusively by parthenogenesis, while others can switch between sexual reproduction and parthenogenesis under certain environmental conditions.
- peak shift model
- peripatric speciation
- A variation of allopatric speciation where a new species forms from a small, peripheral isolated population.[18] It is sometimes referred to as centripetal speciation in contrast to centrifugal speciation.
- phenetic
- Pertaining to phenotypic similarity, e.g. a phenetic classification.[1]
- phenotype
- phyletic gradualism
- phylogenetics
- The study of the evolutionary history and relationships among individuals or groups of organisms (e.g. species or populations within a species).
- phylogenetic bracketing
- phylogenetic species concept
- phylogeny
- phylogeography
- plesiomorphy
- An ancestral character state; i.e. the state or condition of a particular trait or feature (e.g. a specific phenotype) that is present in the common ancestor of a given clade. Plesiomorphies may or may not be shared by some or all descendants within the clade. The term is relative; a trait considered a plesiomorphy in one clade may not be considered a plesiomorphy in a different clade. Contrast apomorphy.
- pollinator isolation
- polymorphism
- polyphyly
- The grouping of organisms which do not share an immediate common ancestor; such groups are said to be polyphyletic. The term is often applied to groups of organisms that share characteristics which appear to be similar but are not actually closely related, frequently as a result of convergent evolution. The avoidance of polyphyletic groupings is often a stimulus for major revisions of biological classification schemes. Contrast monophyly and paraphyly.
- polyploidy
- polytomy
- popular sire effect
- population
- A group of organisms of the same species which occupies a more or less well-defined geographic region and which exhibits reproductive continuity from generation to generation. It is generally presumed that ecological and reproductive interactions occur more frequently among the members of the group than between them and members of other populations of the same species.[1]
- population bottleneck
- A sharp, often sudden reduction in the size of a biological population.
- postmating barrier
- postmating prezygotic isolation
- preadaptation
- Possession of the necessary properties to permit a shift into a new niche or habitat. A structure is said to be preadapted if it can assume a new function before it itself becomes modified by selection.[1]
- premating barrier
- premating isolation
- punctuated equilibrium

A diagram representing population subject to a selective gradient of phenotypic or genotypic frequencies (a cline). Each end of the gradient experiences different selective conditions (divergent selection). Reproductive isolation occurs upon the formation of a hybrid zone. In most cases, the hybrid zone may become eliminated due to a selective disadvantage. This effectively completes the speciation process.

In peripatric speciation, a small population becomes isolated on the periphery of the central population evolving reproductive isolation (blue) due to reduced gene flow
Q[]
- quantum evolution
- A rapid evolutionary shift in a lineage to a phenotypic state that is distinctly unlike the ancestral condition.[1]
- quantum speciation
- A chromosomal model of speciation that occurs rapidly when a cross-fertilizing plant species buds off from a larger population on the periphery, experiencing interbreeding and strong genetic drift that results in a new species.[19][20][21] The model is similar to that of Ernst Mayr's peripatric speciation.[22]
R[]
- r-selection
- reaction norm
- The pattern or set of phenotypic expressions of a given genotype across a variety of different environmental conditions.[1]
- recapitulation
- The ontogenetic passage of an organism's features through stages that resemble the adult features of the organism's phylogenetic ancestors.[1]
- recognition species concept
- recombinational speciation
- recurrent evolution
- The repeated evolution of a particular trait or character, for whatever reason, whether by natural selection or genetic drift.
- Red Queen hypothesis
- refugium
- A geographic location (or, more narrowly, a niche) in which one or more species has persisted while becoming extinct elsewhere.[1]
- reinforcement
- A process of speciation by which natural selection increases the reproductive isolation between two populations of a species as a result of selection acting against the production of hybrid individuals of low fitness.[3] See also Evidence of speciation by reinforcement.
- relict
- A species or population that is the last surviving representative of an otherwise extinct group, taxon, lineage, or clade, or which has been left behind in a locality after extinction throughout most of a formerly larger geographic distribution.[1]
- reproductive character displacement
- reproductive isolating barriers
- The set of mechanisms responsible for speciation.
- reproductive isolation
- When two different species mate and cannot produce fertile offspring. Isolating mechanisms are typically classified as prezygotic (isolating barriers occurring before the formation of a zygote) and postzygotic (isolating barriers occurring after the formation of a zygote).
- reproductive success
- reticulate evolution
- The union of different lineages of a clade by hybridization.[1]
- ring species
- Connected populations of a species, each of which can interbreed with closely sited related populations, but for which there exist at least two "end" populations in the series, which are too distantly related to interbreed.
- robustness
- The persistence of a certain phenotypic trait or characteristic in a biological system despite perturbations or conditions of uncertainty. Robustness is achieved through the combination of many genetic and molecular mechanisms which effectively preserve the integrity of a particular adaptation, and can evolve by direct or indirect selection.
- runaway selection

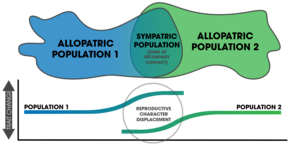
Reproductive character displacement sometimes occurs when two allopatric populations come into secondary contact. Once in sympatry, changes can be seen in mating-associated traits only in the zone of contact. This is a common pattern found in speciation by reinforcement.

In a ring species, individuals are able to successfully reproduce (exchange genes) with members of their own species in adjacent populations occupying a suitable habitat around a geographic barrier. Individuals at the ends of the cline are unable to reproduce when they come into contact.
S[]
- saltation
- A sudden and large mutational change from one generation to the next which is sufficient to cause rapid or immediate speciation. Various forms of saltation, such as by polyploidy in plants, have often historically been interpreted as evidence for certain theories of mutationism, in contrast to Darwinian gradualism.
- secondary contact
- The process by which two allopatrically distributed populations of a species are geographically reunited. Contact between divergent populations may renew the potential for gene flow between them, depending upon how reproductively isolated the populations have become.
- selection
- The non-random differential survival or reproduction of classes of phenotypically different entities.[1] Selection may occur naturally or may be induced artificially. Selection is often studied in different modes (as with sexual selection and kin selection) or from the perspective of distinct units (as with genic selection and group selection).
- selection coefficient
- The difference between the mean relative fitness of individuals of a given genotype and those of a reference genotype.[1]
- selective pressure
- selective sweep
- The process by which strong positive selection of a new and beneficial mutation within a population causes the mutation to reach fixation so quickly that nearby linked DNA sequences also become fixed via genetic hitchhiking, thereby reducing or eliminating the genetic variation of nearby loci within the population.
- semelparity
- A reproductive strategy characterized by a single reproductive episode during an individual organism's lifetime, especially one in which the programmed death of the organism immediately after the reproductive event constitutes part of an overall strategy that includes putting all available resources into maximizing the probability of reproductive success, at the expense of the organism's future life. Organisms that use such a strategy are said to be semelparous. Semelparity is usually contrasted with iteroparity.
- semi-geographic speciation
- semipermeable species boundary
- The idea that gene flow can occur between two species but that certain alleles at particular loci can exchange whereas others cannot.[16] It is often used to describe hybrid zones and has also been referred to as porous.[16]
- semispecies
- One of several groups of populations that are partially but not entirely reproductively isolated from each other by biological isolating mechanisms,[1] and which are therefore neither easily definable as belonging to the same species nor to separate species. The taxon of species itself is not a well-defined concept.
- sexual reproduction
- sexual selection
- spandrel
- speciation
- The evolutionary process by which populations evolve to become distinct species.
- speciation experiment
- An experiment that attempts to replicate reproductive isolation in nature in a scientifically controlled, laboratory setting.
- Speciation that can be detected as occurring in fossilized organisms.
- speciation rate
- species
- A basic unit of biological classification, traditionally interpreted according to the biological species concept as the members in aggregate of a group of populations of organisms which interbreed or potentially interbreed with each other under natural conditions;[1] a basic taxonomic rank to which individual specimens are assigned and which often but not always corresponds to the definition of a biological species; and a fundamental unit used to interpret and measure biodiversity in ecological contexts. The concept of species is notoriously complex and often problematic to define precisely; many different conceptualizations of what is or should be meant by the term have been defined in scientific literature.
- species complex
- species concept
- species problem
- The difficulty in precisely defining what a species is and in determining the placement of an organism within a particular species.[24]
- stasipatric speciation
- stasis
- A species lineage that experiences little phenotypic or genotypic change over time.[7]
- stepping-stone speciation
- sterility
- subspecies
- A named geographic race, or a set of populations of the same species which share one or more distinctive features and occupy an area that is geographically separate from other subspecies.[1] Not all species are formally divided into subspecies, and the taxon of species itself is not a well-defined concept.
- survival of the fittest
- suture zone
- A geographic region that exhibits a significant number of hybrid zones, contact zones between populations, and phylogeographic breaks.[25]
- swamping effect
- sympatric speciation
- sympatry
- symplesiomorphy
- synapomorphy
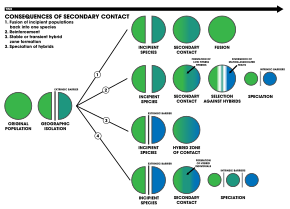
The four outcomes of secondary contact:
1. An extrinsic barrier separates a species population into two but they come into contact before reproductive isolation is sufficient to result in speciation. The two populations fuse back into one species.
2. Speciation by reinforcement.
3. Two separated populations stay genetically distinct while hybrid swarms form in the zone of contact.
4. Genome recombination results in speciation of the two populations, with an additional hybrid species. All three species are separated by intrinsic reproductive barriers.[23]
T[]
U[]
V[]
- vicariance biogeography
- A biogeographic approach to species distributions that uses their phylogenetic histories—patterns resulting from allopatric speciation events in the past.[26]
- vicariant speciation
- A biogeographic term meaning the geographic isolation of two species populations (as in allopatric speciation).
W[]
- Wahlund effect
- A phenomenon by which a reduction of heterozygosity at a particular genetic locus within a population as a whole is observed when two or more subpopulations have different allele frequencies at that locus, even if the subpopulations themselves are each in Hardy–Weinberg equilibrium.
- Wallace effect
Y[]
- Y-chromosomal Adam
See also[]
References[]
- ^ Jump up to: a b c d e f g h i j k l m n o p q r s t u v w x y z aa Futuyma, Douglas J. (1986). Evolutionary biology (2nd ed.). Sunderland, Mass.: Sinauer Associates, Inc. p. 550–556. ISBN 0878931880. Retrieved 15 January 2021.
- ^ Oxford Reference (2008), agamospecies, Oxford University Press
- ^ Jump up to: a b c d e f Jerry A. Coyne; H. Allen Orr (2004), Speciation, Sinauer Associates, pp. 1–545, ISBN 978-0-87893-091-3
- ^ Guy L. Bush (1994), "Sympatric speciation in animals: new wine in old bottles", Trends in Ecology & Evolution, 9 (8): 285–288, doi:10.1016/0169-5347(94)90031-0, PMID 21236856
- ^ Howard, Daniel J. (2003). "Speciation: Allopatric". Encyclopedia of Life Sciences. eLS. doi:10.1038/npg.els.0001748. ISBN 978-0470016176.
- ^ Guy L. Bush (1994), "Sympatric speciation in animals: new wine in old bottles", Trends in Ecology & Evolution, 9 (8): 285–288, doi:10.1016/0169-5347(94)90031-0, PMID 21236856
- ^ Jump up to: a b c d e f g Vaux, Felix; Trewick, Steven A.; Morgan-Richards, Mary (2016). "Lineages, splits and divergence challenge whether the terms anagenesis and cladogenesis are necessary". Biological Journal of the Linnean Society. 117 (2): 165–76. doi:10.1111/bij.12665.
- ^ M. V. Lomolino & J. H. Brown (1998), Biography (2 ed.), Sinauer Associates, Sunderland, MA., pp. 3, ISBN 978-0-87893-073-9
- ^ Sergey Gavrilets; et al. (2000), "Patterns of Parapatric Speciation", Evolution, 54 (4): 1126–1134, doi:10.1554/0014-3820(2000)054[1126:pops]2.0.co;2, PMID 11005282
- ^ Page, Roderick DM. (2006). "Cospeciation". Encyclopedia of Life Sciences. eLS. doi:10.1038/npg.els.0004124. ISBN 978-0470016176.
- ^ Howard D. Rundle and Patrik Nosil (2005), "Ecological Speciation", Ecology Letters, 8 (3): 336–352, doi:10.1111/j.1461-0248.2004.00715.x
- ^ Waters JM, Fraser CI, Hewitt GM (2013). "Founder takes all: density-dependent processes structure biodiversity". Trends in Ecology & Evolution. 28 (2): 78–85. doi:10.1016/j.tree.2012.08.024. PMID 23000431.
- ^ Turelli, M; Orr, H.A. (May 1995). "The Dominance Theory of Haldane's Rule". Genetics. 140 (1): 389–402. PMC 1206564. PMID 7635302.
- ^ Anders Ödeen & Ann-Britt Florin (2002), "Sexual selection and peripatric speciation: the Kaneshiro model revisited", Journal of Evolutionary Biology, 15 (2): 301–306, doi:10.1046/j.1420-9101.2002.00378.x, S2CID 82095639
- ^ B. M. Fitzpatrick; A. A. Fordyce; S. Gavrilets (2008), "What, if anything, is sympatric speciation?", Journal of Evolutionary Biology, 21 (6): 1452–1459, doi:10.1111/j.1420-9101.2008.01611.x, PMID 18823452, S2CID 8721116
- ^ Jump up to: a b c d e f Richard G. Harrison (2012), "The Language of Speciation", Evolution, 66 (12): 3643–3657, doi:10.1111/j.1558-5646.2012.01785.x, PMID 23206125, S2CID 31893065
- ^ B. B. Fitzpatrick, J. A. Fordyce, & S. Gavrilets (2009), "Pattern, process and geographic modes of speciation", Journal of Evolutionary Biology, 22 (11): 2342–2347, doi:10.1111/j.1420-9101.2009.01833.x, PMID 19732257, S2CID 941124CS1 maint: multiple names: authors list (link)
- ^ Michael Turelli, Nicholas H. Barton, and Jerry A. Coyne (2001), "Theory and speciation", Trends in Ecology & Evolution, 16 (7): 330–343, doi:10.1016/s0169-5347(01)02177-2, PMID 11403865CS1 maint: multiple names: authors list (link)
- ^ Verne Grant (1971), Plant Speciation, New York: Columbia University Press, p. 432, ISBN 978-0231083263
- ^ Douglas J. Futuyma (1989), "Speciational trends and the role of species in macroevolution", The American Naturalist, 134 (2): 318–321, doi:10.1086/284983
- ^ Loren H. Rieseberg (2001), "Chromosomal rearrangements and speciation", Trends in Ecology & Evolution, 16 (7): 351–358, doi:10.1016/s0169-5347(01)02187-5, PMID 11403867
- ^ L. D. Gottlieb (2003), "Rethinking classic examples of recent speciation in plants", New Phytologist, 161: 71–82, doi:10.1046/j.1469-8137.2003.00922.x
- ^ Hvala, John A.; Wood, Troy E. (2012). Speciation: Introduction. eLS. doi:10.1002/9780470015902.a0001709.pub3. ISBN 978-0470016176.
- ^ William P. Hanage (2013), "Fuzzy species revisited", BMC Biology, 11 (41): 41, doi:10.1186/1741-7007-11-41, PMC 3626887, PMID 23587266
- ^ Luciano Nicolas Naka, Catherine L. Bechtoldt, L. Magalli Pinto Henriques, and Robb T. Brumfield (2012), "The Role of Physical Barriers in the Location of Avian Suture Zones in the Guiana Shield, Northern Amazonia", The American Naturalist, 179 (4), doi:10.1086/664627CS1 maint: multiple names: authors list (link)
- ^ M. V. Lomolino & J. H. Brown (1998), Biography (2 ed.), Sinauer Associates, Sunderland, MA., pp. 352–357, ISBN 978-0-87893-073-9
Categories:
- Evolutionary biology
- Speciation
- Glossaries of biology
