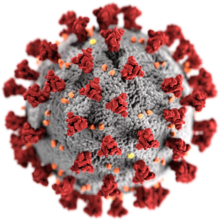
Severe acute respiratory syndrome–related coronavirus
| Severe acute respiratory syndrome–related coronavirus | |
|---|---|

| |
| Transmission electron micrograph of SARS-related coronaviruses emerging from host cells cultured in the lab | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Sarbecovirus |
| Species: | Severe acute respiratory syndrome–related coronavirus
|
| Strains | |
| |
| Synonyms | |
| |
Severe acute respiratory syndrome–related coronavirus (SARSr-CoV or SARS-CoV)[note 1] is a species of virus consisting of many known strains phylogenetically related to severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) that have been shown to possess the capability to infect humans, bats, and certain other mammals.[2][3] These enveloped, positive-sense single-stranded RNA viruses enter host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor.[4] The SARSr-CoV species is a member of the genus Betacoronavirus and of the subgenus Sarbecovirus.[5][6]
Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1 (SARS-CoV or SARS-CoV-1), which caused the 2002–2004 outbreak of severe acute respiratory syndrome (SARS), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is causing the ongoing pandemic of COVID-19.[7][8] There are hundreds of other strains of SARSr-CoV, which are only known to infect non-human species: bats are a major reservoir of many strains of SARSr-CoV; several strains have been identified in palm civets, which were likely ancestors of SARS-CoV-1.[7][9]
The SARS-related coronavirus was one of several viruses identified by the World Health Organization (WHO) in 2016 as a likely cause of a future epidemic in a new plan developed after the Ebola epidemic for urgent research and development before and during an epidemic towards diagnostic tests, vaccines and medicines. This prediction came to pass in 2020 with the COVID-19 pandemic.[10][11]
Classification[]
SARS-related coronavirus is a member of the genus Betacoronavirus (group 2) and monotypic of the subgenus Sarbecovirus (subgroup B).[12] Sarbecoviruses, unlike embecoviruses or alphacoronaviruses, have only one () instead of two in the open reading frame ORF1.[13] SARSr-CoV was determined to be an early split-off from the betacoronaviruses based on a set of conserved domains that it shares with the group.[14][15]
Bats serve as the main host reservoir species for the SARS-related coronaviruses like SARS-CoV-1 and SARS-CoV-2. The virus has coevolved in the bat host reservoir over a long period of time.[16] Only recently have strains of SARS-related coronavirus been observed to have evolved into having been able to make the cross-species jump from bats to humans, as in the case of the strains SARS-CoV and SARS-CoV-2.[17][4] Both of these strains descended from a single ancestor but made the cross-species jump into humans separately. SARS-CoV-2 is not a direct descendant of SARS-CoV.[7]
Genome[]

The SARS-related coronavirus is an enveloped, positive-sense, single-stranded RNA virus. Its genome is about 30 kb, which is one of the largest among RNA viruses. The virus has 14 open reading frames which overlap in some cases.[18] The genome has the usual 5′ methylated cap and a 3′ polyadenylated tail.[19] There are 265 nucleotides in the 5'UTR and 342 nucleotides in the 3'UTR.[18]
The 5' methylated cap and 3' polyadenylated tail allows the positive-sense RNA genome to be directly translated by the host cell's ribosome on viral entry.[20] SARSr-CoV is similar to other coronaviruses in that its genome expression starts with translation by the host cell's ribosomes of its initial two large overlapping open reading frames (ORFs), 1a and 1b, both of which produce polyproteins.[18]
| showFunction of SARS-CoV genome proteins (orf1a to orf9b) |
|---|
The functions of several of the viral proteins are known.[24] ORFs 1a and 1b encode the replicase/transcriptase polyprotein, and later ORFs 2, 4, 5, and 9a encode, respectively, the four major structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N).[25] The later ORFs also encode for eight unique proteins (orf3a to orf9b), known as the accessory proteins, many with no known homologues. The different functions of the accessory proteins are not well understood.[24]
SARS coronaviruses have been genetically engineered in several laboratories.[26]
Phylogenetics[]
Phylogenetic analysis showed that the evolutionary branch composed of Bat coronavirus BtKY72 and BM48-31 was the base group of SARS–related CoVs evolutionary tree, which separated from other SARS–related CoVs earlier than SARS-CoV-1 and SARS-CoV-2.[27][28]
| SARSr‑CoV |
| ||||||||||||||||||||||||||||||||||||||||||
[]
A phylogenetic tree based on whole-genome sequences of SARS-CoV-1 and related coronaviruses is:
| SARS‑CoV‑1 related coronavirus |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
SARS-CoV-2, 79% to SARS-CoV-1[36] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
[]
A phylogenetic tree based on whole-genome sequences of SARS-CoV-2 and related coronaviruses is:[37][38]
| SARS‑CoV‑2 related coronavirus |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
SARS-CoV-1, 79% to SARS-CoV-2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Morphology[]

The morphology of the SARS-related coronavirus is characteristic of the coronavirus family as a whole. The viruses are large pleomorphic spherical particles with bulbous surface projections that form a corona around the particles in electron micrographs.[46] The size of the virus particles is in the 80–90 nm range. The envelope of the virus in electron micrographs appears as a distinct pair of electron dense shells.[47]
The viral envelope consists of a lipid bilayer where the membrane (M), envelope (E) and spike (S) proteins are anchored.[48] The spike proteins provide the virus with its bulbous surface projections. The spike protein's interaction with its complement host cell receptor is central in determining the tissue tropism, infectivity, and species range of the virus.[49][50]
Inside the envelope, there is the nucleocapsid, which is formed from multiple copies of the nucleocapsid (N) protein, which are bound to the positive-sense single-stranded (~30 kb) RNA genome in a continuous beads-on-a-string type conformation.[51][52] The lipid bilayer envelope, membrane proteins, and nucleocapsid protect the virus when it is outside the host.[53]
Life cycle[]
SARS-related coronavirus follows the replication strategy typical of all coronaviruses.[19][54]
Attachment and entry[]

The attachment of the SARS-related coronavirus to the host cell is mediated by the spike protein and its receptor.[55] The spike protein receptor binding domain (RBD) recognizes and attaches to the angiotensin-converting enzyme 2 (ACE2) receptor.[4] Following attachment, the virus can enter the host cell by two different paths. The path the virus takes depends on the host protease available to cleave and activate the receptor-attached spike protein.[56]
The first path the SARS coronavirus can take to enter the host cell is by endocytosis and uptake of the virus in an endosome. The receptor-attached spike protein is then activated by the host's pH-dependent cysteine protease cathepsin L. Activation of the receptor-attached spike protein causes a conformational change, and the subsequent fusion of the viral envelope with the endosomal wall.[56]
Alternatively, the virus can enter the host cell directly by proteolytic cleavage of the receptor-attached spike protein by the host's TMPRSS2 or TMPRSS11D serine proteases at the cell surface.[57][58] In the SARS coronavirus, the activation of the C-terminal part of the spike protein triggers the fusion of the viral envelope with the host cell membrane by inducing conformational changes which are not fully understood.[59]
Genome translation[]
| showFunction of coronavirus nonstructural proteins (nsps)[60] |
|---|
After fusion the nucleocapsid passes into the cytoplasm, where the viral genome is released.[55] The genome acts as a messenger RNA, and the cell's ribosome translates two-thirds of the genome, which corresponds to the open reading frame ORF1a and ORF1b, into two large overlapping polyproteins, pp1a and pp1ab.
The larger polyprotein pp1ab is a result of a -1 ribosomal frameshift caused by a slippery sequence (UUUAAAC) and a downstream RNA pseudoknot at the end of open reading frame ORF1a.[61] The ribosomal frameshift allows for the continuous translation of ORF1a followed by ORF1b.[62]
The polyproteins contain their own proteases, PLpro and 3CLpro, which cleave the polyproteins at different specific sites. The cleavage of polyprotein pp1ab yields 16 nonstructural proteins (nsp1 to nsp16). Product proteins include various replication proteins such as RNA-dependent RNA polymerase (RdRp), RNA helicase, and exoribonuclease (ExoN).[62]
The two SARS-CoV-2 proteases (PLpro and 3CLpro) also interfere with the immune system response to the viral infection by cleaving three immune system proteins. PLpro cleaves IRF3 and 3CLpro cleaves both NLRP12 and TAB1. "Direct cleavage of IRF3 by NSP3 could explain the blunted Type-I IFN response seen during SARS-CoV-2 infections while NSP5 mediated cleavage of NLRP12 and TAB1 point to a molecular mechanism for enhanced production of IL-6 and inflammatory response observed in COVID-19 patients."[63]
Replication and transcription[]

A number of the nonstructural replication proteins coalesce to form a multi-protein replicase-transcriptase complex (RTC).[62] The main replicase-transcriptase protein is the RNA-dependent RNA polymerase (RdRp). It is directly involved in the replication and transcription of RNA from an RNA strand. The other nonstructural proteins in the complex assist in the replication and transcription process.[60]
The protein nsp14 is a 3'-5' exoribonuclease which provides extra fidelity to the replication process. The exoribonuclease provides a proofreading function to the complex which the RNA-dependent RNA polymerase lacks. Similarly, proteins nsp7 and nsp8 form a hexadecameric sliding clamp as part of the complex which greatly increases the processivity of the RNA-dependent RNA polymerase.[60] The coronaviruses require the increased fidelity and processivity during RNA synthesis because of the relatively large genome size in comparison to other RNA viruses.[64]
One of the main functions of the replicase-transcriptase complex is to transcribe the viral genome. RdRp directly mediates the synthesis of negative-sense subgenomic RNA molecules from the positive-sense genomic RNA. This is followed by the transcription of these negative-sense subgenomic RNA molecules to their corresponding positive-sense mRNAs.[65]
The other important function of the replicase-transcriptase complex is to replicate the viral genome. RdRp directly mediates the synthesis of negative-sense genomic RNA from the positive-sense genomic RNA. This is followed by the replication of positive-sense genomic RNA from the negative-sense genomic RNA.[65]
The replicated positive-sense genomic RNA becomes the genome of the progeny viruses. The various smaller mRNAs are transcripts from the last third of the virus genome which follows the reading frames ORF1a and ORF1b. These mRNAs are translated into the four structural proteins (S, E, M, and N) that will become part of the progeny virus particles and also eight other accessory proteins (orf3 to orf9b) which assist the virus.[66]
Recombination[]
When two SARS-CoV genomes are present in a host cell, they may interact with each other to form recombinant genomes that can be transmitted to progeny viruses. Recombination likely occurs during genome replication when the RNA polymerase switches from one template to another (copy choice recombination).[67] Human SARS-CoV appears to have had a complex history of recombination between ancestral coronaviruses that were hosted in several different animal groups.[67][68]
Assembly and release[]
RNA translation occurs inside the endoplasmic reticulum. The viral structural proteins S, E and M move along the secretory pathway into the Golgi intermediate compartment. There, the M proteins direct most protein-protein interactions required for assembly of viruses following its binding to the nucleocapsid.[69]
Progeny viruses are released from the host cell by exocytosis through secretory vesicles.[69]
See also[]
- Bat SARS-like coronavirus WIV1 (SL-CoV-WIV1)
- Bat SARS-like coronavirus RsSHC014
- Bat coronavirus RaTG13
- Civet SARS-CoV
Notes[]
- ^ The terms SARSr-CoV and SARS-CoV are sometimes used interchangeably, especially prior to the discovery of SARS-CoV-2. This may cause confusion when some publications refer to SARS-CoV-1 as SARS-CoV.
References[]
- ^ "ICTV Taxonomy history: Severe acute respiratory syndrome-related coronavirus". International Committee on Taxonomy of Viruses (ICTV). Retrieved 27 January 2019.
- ^ Branswell H (9 November 2015). "SARS-like virus in bats shows potential to infect humans, study finds". Stat News. Retrieved 20 February 2020.
- ^ Wong AC, Li X, Lau SK, Woo PC (February 2019). "Global Epidemiology of Bat Coronaviruses". Viruses. 11 (2): 174. doi:10.3390/v11020174. PMC 6409556. PMID 30791586.
Most notably, horseshoe bats were found to be the reservoir of SARS-like CoVs, while palm civet cats are considered to be the intermediate host for SARS-CoVs [43,44,45].
- ^ Jump up to: a b c Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. (November 2013). "Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor". Nature. 503 (7477): 535–8. Bibcode:2013Natur.503..535G. doi:10.1038/nature12711. PMC 5389864. PMID 24172901.
- ^ "Virus Taxonomy: 2018 Release". International Committee on Taxonomy of Viruses (ICTV). October 2018. Retrieved 13 January 2019.
- ^ Woo PC, Huang Y, Lau SK, Yuen KY (August 2010). "Coronavirus genomics and bioinformatics analysis". Viruses. 2 (8): 1804–20. doi:10.3390/v2081803. PMC 3185738. PMID 21994708.
Figure 2. Phylogenetic analysis of RNA-dependent RNA polymerases (Pol) of coronaviruses with complete genome sequences available. The tree was constructed by the neighbor-joining method and rooted using Breda virus polyprotein.
- ^ Jump up to: a b c Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (March 2020). "The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2". Nature Microbiology. 5 (4): 536–544. doi:10.1038/s41564-020-0695-z. PMC 7095448. PMID 32123347.
- ^ Kohen, Jon; Kupferschmidth, Kai (28 February 2020). "Strategies shift as coronavirus pandemic looms". Science. 367 (6481): 962–963. Bibcode:2020Sci...367..962C. doi:10.1126/science.367.6481.962. PMID 32108093.
- ^ Lau SK, Li KS, Huang Y, Shek CT, Tse H, Wang M, et al. (March 2010). "Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events". Journal of Virology. 84 (6): 2808–19. doi:10.1128/JVI.02219-09. PMC 2826035. PMID 20071579.
- ^ Kieny M. "After Ebola, a Blueprint Emerges to Jump-Start R&D". Scientific American Blog Network. Archived from the original on 20 December 2016. Retrieved 13 December 2016.
- ^ "LIST OF PATHOGENS". World Health Organization. Archived from the original on 20 December 2016. Retrieved 13 December 2016.
- ^ Wong AC, Li X, Lau SK, Woo PC (February 2019). "Global Epidemiology of Bat Coronaviruses". Viruses. 11 (2): 174. doi:10.3390/v11020174. PMC 6409556. PMID 30791586.
See Figure 1.
- ^ Woo PC, Huang Y, Lau SK, Yuen KY (August 2010). "Coronavirus genomics and bioinformatics analysis". Viruses. 2 (8): 1804–20. doi:10.3390/v2081803. PMC 3185738. PMID 21994708.
See Figure 1.
- ^ Woo PC, Huang Y, Lau SK, Yuen KY (August 2010). "Coronavirus genomics and bioinformatics analysis". Viruses. 2 (8): 1804–20. doi:10.3390/v2081803. PMC 3185738. PMID 21994708.
Furthermore, subsequent phylogenetic analysis using both complete genome sequence and proteomic approaches, it was concluded that SARSr-CoV is probably an early split-off from the Betacoronavirus lineage [1]; See Figure 2.
- ^ "Coronaviridae - Figures - Positive Sense RNA Viruses - Positive Sense RNA Viruses (2011)". International Committee on Taxonomy of Viruses (ICTV). Retrieved 6 March 2020.
See Figure 2.
- ^ Gouilh MA, Puechmaille SJ, Gonzalez JP, Teeling E, Kittayapong P, Manuguerra JC (October 2011). "SARS-Coronavirus ancestor's foot-prints in South-East Asian bat colonies and the refuge theory". Infection, Genetics and Evolution. 11 (7): 1690–702. doi:10.1016/j.meegid.2011.06.021. PMC 7106191. PMID 21763784.
Betacoronaviruses-b ancestors, meaning SARSr-CoVs ancestors, could have been historically hosted by the common ancestor of the Rhinolophidae and Hipposideridae and could have later evolved independently in the lineages leading towards Rhinolophidae and Hipposideridae betacoronaviruses.
- ^ Cui J, Han N, Streicker D, Li G, Tang X, Shi Z, et al. (October 2007). "Evolutionary relationships between bat coronaviruses and their hosts". Emerging Infectious Diseases. 13 (10): 1526–32. doi:10.3201/eid1310.070448. PMC 2851503. PMID 18258002.
- ^ Jump up to: a b c Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. (August 2003). "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". Journal of Molecular Biology. 331 (5): 991–1004. doi:10.1016/S0022-2836(03)00865-9. PMC 7159028. PMID 12927536.
The SARS-CoV genome is ∼29.7 kb long and contains 14 open reading frames (ORFs) flanked by 5′ and 3′-untranslated regions of 265 and 342 nucleotides, respectively (Figure 1).
- ^ Jump up to: a b Fehr AR, Perlman S (2015). "Coronaviruses: an overview of their replication and pathogenesis". In Maier HJ, Bickerton E, Britton P (eds.). Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
- ^ Fehr AR, Perlman S (2015). Maier HJ, Bickerton E, Britton P (eds.). An Overview of Their Replication and Pathogenesis; Section 2 Genomic Organization. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
- ^ McBride R, Fielding BC (November 2012). "The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis". Viruses. 4 (11): 2902–23. doi:10.3390/v4112902. PMC 3509677. PMID 23202509.
See Table 1.
- ^ Tang X, Li G, Vasilakis N, Zhang Y, Shi Z, Zhong Y, Wang LF, Zhang S (March 2009). "Differential stepwise evolution of SARS coronavirus functional proteins in different host species". BMC Evolutionary Biology. 9: 52. doi:10.1186/1471-2148-9-52. PMC 2676248. PMID 19261195.
- ^ Narayanan, Krishna; Huang, Cheng; Makino, Shinji (April 2008). "SARS coronavirus Accessory Proteins". Virus Research. 133 (1): 113–121. doi:10.1016/j.virusres.2007.10.009. ISSN 0168-1702. PMC 2720074. PMID 18045721.
See Table 1.
- ^ Jump up to: a b McBride R, Fielding BC (November 2012). "The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis". Viruses. 4 (11): 2902–23. doi:10.3390/v4112902. PMC 3509677. PMID 23202509.
- ^ Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. (August 2003). "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". Journal of Molecular Biology. 331 (5): 991–1004. doi:10.1016/S0022-2836(03)00865-9. PMC 7159028. PMID 12927536.
See Figure 1.
- ^ Kaina, Bernd (2021). "On the Origin of SARS-CoV-2: Did Cell Culture Experiments Lead to Increased Virulence of the Progenitor Virus for Humans?". In Vivo. 35 (3): 1313–1326. doi:10.21873/invivo.12384. PMC 8193286. PMID 33910809.
- ^ Lu R, Zhao X, Li J, Niu P, Yang B, Wu H; et al. (2020). "Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding". Lancet. 395 (10224): 565–574. doi:10.1016/S0140-6736(20)30251-8. PMC 7159086. PMID 32007145.CS1 maint: multiple names: authors list (link)
- ^ Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020). "The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2". Nat Microbiol. 5 (4): 536–544. doi:10.1038/s41564-020-0695-z. PMC 7095448. PMID 32123347.
- ^ Kim Y, Son K, Kim YS, Lee SY, Jheong W, Oem JK (2019). "Complete genome analysis of a SARS-like bat coronavirus identified in the Republic of Korea". Virus Genes. 55 (4): 545–549. doi:10.1007/s11262-019-01668-w. PMC 7089380. PMID 31076983.CS1 maint: multiple names: authors list (link)
- ^ Jump up to: a b Li, W. (2005). "Bats Are Natural Reservoirs of SARS-Like Coronaviruses". Science. 310 (5748): 676–679. doi:10.1126/science.1118391. ISSN 0036-8075.
- ^ Jump up to: a b Xing‐Yi Ge, Ben Hu, and Zheng‐Li Shi (2015). "BAT CORONAVIRUSES". In Lin-Fa Wang and Christopher Cowled (ed.). Bats and Viruses: A New Frontier of Emerging Infectious Diseases, First Edition. John Wiley & Sons.CS1 maint: multiple names: authors list (link)
- ^ He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y; et al. (2014). "Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China". J Virol. 88 (12): 7070–82. doi:10.1128/JVI.00631-14. PMC 4054348. PMID 24719429.CS1 maint: multiple names: authors list (link)
- ^ Jump up to: a b Lau, Susanna K. P.; Feng, Yun; Chen, Honglin; Luk, Hayes K. H.; Yang, Wei-Hong; Li, Kenneth S. M.; Zhang, Yu-Zhen; Huang, Yi; et al. (2015). "Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination". Journal of Virology. 89 (20): 10532–10547. doi:10.1128/JVI.01048-15. ISSN 0022-538X.
- ^ Jump up to: a b Xing-Yi Ge; Jia-Lu Li; Xing-Lou Yang; et al. (2013). "Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor". Nature. 503 (7477): 535–8. Bibcode:2013Natur.503..535G. doi:10.1038/nature12711. PMC 5389864. PMID 24172901.
- ^ Yang XL, Hu B, Wang B, Wang MN, Zhang Q, Zhang W; et al. (2016). "Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus". J Virol. 90 (6): 3253–6. doi:10.1128/JVI.02582-15. PMC 4810638. PMID 26719272.CS1 maint: multiple names: authors list (link)
- ^ Ben, Hu; Hua, Guo; Peng, Zhou; Zheng-Li, Shi (2020). "Characteristics of SARS-CoV-2 and COVID-19". Nature Reviews Microbiology (19): 141–154. doi:10.1038/s41579-020-00459-7.
- ^ Jump up to: a b Zhou H, Ji J, Chen X, Bi Y, Li J, Wang Q, et al. (June 2021). "Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses". Cell. 184 (17): 4380–4391.e14. doi:10.1016/j.cell.2021.06.008. PMC 8188299. PMID 34147139.
- ^ Jump up to: a b Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, et al. (February 2021). "Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia". Nature Communications. 12 (1): 972. Bibcode:2021NatCo..12..972W. doi:10.1038/s41467-021-21240-1. PMC 7873279. PMID 33563978.
- ^ Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, et al. (December 2020). "Detection and Characterization of Bat Sarbecovirus Phylogenetically Related to SARS-CoV-2, Japan". Emerging Infectious Diseases. 26 (12): 3025–3029. doi:10.3201/eid2612.203386. PMC 7706965. PMID 33219796.
- ^ Jump up to: a b Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, et al. (June 2020). "A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein". Current Biology. 30 (11): 2196–2203.e3. doi:10.1016/j.cub.2020.05.023. PMC 7211627. PMID 32416074.
- ^ Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, et al. (July 2020). "Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins". Nature. 583 (7815): 282–285. Bibcode:2020Natur.583..282L. doi:10.1038/s41586-020-2169-0. PMID 32218527. S2CID 214683303.
- ^ Liu P, Jiang JZ, Wan XF, Hua Y, Li L, Zhou J, et al. (May 2020). "Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)?". PLOS Pathogens. 16 (5): e1008421. doi:10.1371/journal.ppat.1008421. PMC 7224457. PMID 32407364.
- ^ Jump up to: a b Hul V, Delaune D, Karlsson EA, Hassanin A, Tey PO, Baidaliuk A, et al. (26 January 2021). "A novel SARS-CoV-2 related coronavirus in bats from Cambodia". bioRxiv 10.1101/2021.01.26.428212.
- ^ Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, et al. (June 2020). "A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein". Current Biology. 30 (11): 2196–2203.e3. doi:10.1016/j.cub.2020.05.023. PMC 7211627. PMID 32416074.
- ^ Sonnevend, Julia (December 2020). Alexander, Jeffrey C.; Jacobs, Ronald N.; Smith, Philip (eds.). "A virus as an icon: the 2020 pandemic in images" (PDF). American Journal of Cultural Sociology. Basingstoke: Palgrave Macmillan. 8 (3: The COVID Crisis and Cultural Sociology: Alone Together): 451–461. doi:10.1057/s41290-020-00118-7. eISSN 2049-7121. ISSN 2049-7113.
- ^ Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, et al. (February 2004). "Ultrastructural characterization of SARS coronavirus". Emerging Infectious Diseases. 10 (2): 320–6. doi:10.3201/eid1002.030913. PMC 3322934. PMID 15030705.
Virions acquired an envelope by budding into the cisternae and formed mostly spherical, sometimes pleomorphic, particles that averaged 78 nm in diameter (Figure 1A).
- ^ Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, et al. (August 2006). "Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy". Journal of Virology. 80 (16): 7918–28. doi:10.1128/JVI.00645-06. PMC 1563832. PMID 16873249.
Particle diameters ranged from 50 to 150 nm, excluding the spikes, with mean particle diameters of 82 to 94 nm; Also See Figure 1 for double shell.
- ^ Lai MM, Cavanagh D (1997). "The molecular biology of coronaviruses". Advances in Virus Research. 48: 1–100. doi:10.1016/S0065-3527(08)60286-9. ISBN 9780120398485. PMC 7130985. PMID 9233431.
- ^ Masters PS (1 January 2006). The molecular biology of coronaviruses. Advances in Virus Research. 66. Academic Press. pp. 193–292. doi:10.1016/S0065-3527(06)66005-3. ISBN 9780120398690. PMC 7112330. PMID 16877062.
Nevertheless, the interaction between S protein and receptor remains the principal, if not sole, determinant of coronavirus host species range and tissue tropism.
- ^ Cui J, Li F, Shi ZL (March 2019). "Origin and evolution of pathogenic coronaviruses". Nature Reviews. Microbiology. 17 (3): 181–192. doi:10.1038/s41579-018-0118-9. PMC 7097006. PMID 30531947.
Different SARS-CoV strains isolated from several hosts vary in their binding affinities for human ACE2 and consequently in their infectivity of human cells76,78 (Fig. 6b)
- ^ Fehr AR, Perlman S (2015). Maier HJ, Bickerton E, Britton P (eds.). An Overview of Their Replication and Pathogenesis; Section 2 Genomic Organization. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See section: Virion Structure.
- ^ Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH (March 2014). "The SARS coronavirus nucleocapsid protein--forms and functions". Antiviral Research. 103: 39–50. doi:10.1016/j.antiviral.2013.12.009. PMC 7113676. PMID 24418573.
See Figure 4c.
- ^ Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, et al. (April 2011). "A structural analysis of M protein in coronavirus assembly and morphology". Journal of Structural Biology. 174 (1): 11–22. doi:10.1016/j.jsb.2010.11.021. PMC 4486061. PMID 21130884.
See Figure 10.
- ^ Lal SK, ed. (2010). Molecular Biology of the SARS-Coronavirus. doi:10.1007/978-3-642-03683-5. ISBN 978-3-642-03682-8.
- ^ Jump up to: a b Fehr AR, Perlman S (2015). "Coronaviruses: an overview of their replication and pathogenesis". In Maier HJ, Bickerton E, Britton P (eds.). Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See section: Coronavirus Life Cycle – Attachment and Entry
- ^ Jump up to: a b Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S (December 2013). "Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research". Antiviral Research. 100 (3): 605–14. doi:10.1016/j.antiviral.2013.09.028. PMC 3889862. PMID 24121034.
See Figure 2.
- ^ Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S (January 2014). "TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein". Journal of Virology. 88 (2): 1293–307. doi:10.1128/JVI.02202-13. PMC 3911672. PMID 24227843.
The SARS-CoV can hijack two cellular proteolytic systems to ensure the adequate processing of its S protein. Cleavage of SARS-S can be facilitated by cathepsin L, a pH-dependent endo-/lysosomal host cell protease, upon uptake of virions into target cell endosomes (25). Alternatively, the type II transmembrane serine proteases (TTSPs) TMPRSS2 and HAT can activate SARS-S, presumably by cleavage of SARS-S at or close to the cell surface, and activation of SARS-S by TMPRSS2 allows for cathepsin L-independent cellular entry (26,–28).
- ^ Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY (May 2016). "Coronaviruses - drug discovery and therapeutic options". Nature Reviews. Drug Discovery. 15 (5): 327–47. doi:10.1038/nrd.2015.37. PMC 7097181. PMID 26868298.
S is activated and cleaved into the S1 and S2 subunits by other host proteases, such as transmembrane protease serine 2 (TMPRSS2) and TMPRSS11D, which enables cell surface non-endosomal virus entry at the plasma membrane.
- ^ Li Z, Tomlinson AC, Wong AH, Zhou D, Desforges M, Talbot PJ, et al. (October 2019). "The human coronavirus HCoV-229E S-protein structure and receptor binding". eLife. 8. doi:10.7554/eLife.51230. PMC 6970540. PMID 31650956.
- ^ Jump up to: a b c Fehr AR, Perlman S (2015). "Coronaviruses: an overview of their replication and pathogenesis". In Maier HJ, Bickerton E, Britton P (eds.). Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See Table 2.
- ^ Masters PS (1 January 2006). "The molecular biology of coronaviruses". Advances in Virus Research. Academic Press. 66: 193–292. doi:10.1016/S0065-3527(06)66005-3. ISBN 9780120398690. PMC 7112330. PMID 16877062.
See Figure 8.
- ^ Jump up to: a b c Fehr AR, Perlman S (2015). "Coronaviruses: an overview of their replication and pathogenesis". In Maier HJ, Bickerton E, Britton P (eds.). Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See section: Replicase Protein Expression
- ^ Mehdi Moustaqil (5 June 2020). "SARS-CoV-2 proteases cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species and the search for reservoir hosts". bioRxiv: 2020.06.05.135699. doi:10.1101/2020.06.05.135699. S2CID 219604020.
- ^ Sexton NR, Smith EC, Blanc H, Vignuzzi M, Peersen OB, Denison MR (August 2016). "Homology-Based Identification of a Mutation in the Coronavirus RNA-Dependent RNA Polymerase That Confers Resistance to Multiple Mutagens". Journal of Virology. 90 (16): 7415–28. doi:10.1128/JVI.00080-16. PMC 4984655. PMID 27279608.
Finally, these results, combined with those from previous work (33, 44), suggest that CoVs encode at least three proteins involved in fidelity (nsp12-RdRp, nsp14-ExoN, and nsp10), supporting the assembly of a multiprotein replicase-fidelity complex, as described previously (38).
- ^ Jump up to: a b Fehr AR, Perlman S (2015). "Coronaviruses: an overview of their replication and pathogenesis". In Maier HJ, Bickerton E, Britton P (eds.). Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See section: Corona Life Cycle – Replication and Transcription
- ^ Fehr AR, Perlman S (2015). "Coronaviruses: an overview of their replication and pathogenesis". In Maier HJ, Bickerton E, Britton P (eds.). Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See Figure 1.
- ^ Jump up to: a b Zhang XW, Yap YL, Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch Virol. 2005 Jan;150(1):1-20. Epub 2004 Oct 11. PMID 15480857
- ^ Stanhope MJ, Brown JR, Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect Genet Evol. 2004 Mar;4(1):15-9. PMID 15019585
- ^ Jump up to: a b Fehr AR, Perlman S (2015). "Coronaviruses: an overview of their replication and pathogenesis". In Maier HJ, Bickerton E, Britton P (eds.). Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See section: Coronavirus Life Cycle – Assembly and Release
Further reading[]
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. (April 2003). "Coronavirus as a possible cause of severe acute respiratory syndrome". Lancet. 361 (9366): 1319–25. doi:10.1016/s0140-6736(03)13077-2. PMC 7112372. PMID 12711465.
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. (May 2003). "Characterization of a novel coronavirus associated with severe acute respiratory syndrome". Science. 300 (5624): 1394–9. Bibcode:2003Sci...300.1394R. doi:10.1126/science.1085952. PMID 12730500.
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, et al. (May 2003). "The Genome sequence of the SARS-associated coronavirus". Science. 300 (5624): 1399–404. Bibcode:2003Sci...300.1399M. doi:10.1126/science.1085953. PMID 12730501.
- Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. (August 2003). "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". Journal of Molecular Biology. 331 (5): 991–1004. CiteSeerX 10.1.1.319.7007. doi:10.1016/S0022-2836(03)00865-9. PMC 7159028. PMID 12927536. S2CID 14974326.
- Yount B, Roberts RS, Lindesmith L, Baric RS (August 2006). "Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome". Proceedings of the National Academy of Sciences of the United States of America. 103 (33): 12546–51. Bibcode:2006PNAS..10312546Y. doi:10.1073/pnas.0605438103. PMC 1531645. PMID 16891412.
- Thiel V, ed. (2007). Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-16-5.
- Enjuanes L, Sola I, Zúñiga S, Almazán F (2008). "Coronavirus Replication and Interaction with Host". In Mettenleiter TC, Sobrino F (eds.). Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6.
External links[]
 Media related to Severe acute respiratory syndrome-related coronavirus at Wikimedia Commons
Media related to Severe acute respiratory syndrome-related coronavirus at Wikimedia Commons Data related to Severe acute respiratory syndrome–related coronavirus at Wikispecies
Data related to Severe acute respiratory syndrome–related coronavirus at Wikispecies- WHO press release identifying and naming the SARS virus
- The SARS virus genetic map
- Science special on the SARS virus (free content: no registration required)
- McGill University SARS Resources at the Wayback Machine (archived 1 March 2005)
- U.S. Centers for Disease Control and Prevention (CDC) SARS home
- World Health Organization on alert
- Sarbecovirus